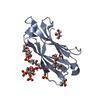
Entry Database : PDB / ID : 6jckTitle Complex structure of Axin-DIX and Dvl2-DIX Axin-1 Segment polarity protein dishevelled homolog DVL-2 Keywords / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Homo sapiens (human)Method / / / Resolution : 3.09 Å Authors Yamanishi, K. / Shibata, N. Funding support Organization Grant number Country Ministry of Education, Culture, Sports, Science and Technology (Japan) 23121526 Ministry of Education, Culture, Sports, Science and Technology (Japan) 25121731
Journal : Sci.Signal. / Year : 2019Title : A direct heterotypic interaction between the DIX domains of Dishevelled and Axin mediates signaling to beta-catenin.Authors : Yamanishi, K. / Fiedler, M. / Terawaki, S.I. / Higuchi, Y. / Bienz, M. / Shibata, N. History Deposition Jan 29, 2019 Deposition site / Processing site Revision 1.0 Jan 15, 2020 Provider / Type Revision 1.1 Nov 22, 2023 Group / Database references / Refinement descriptionCategory chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model Item / _database_2.pdbx_database_accession
Show all Show less
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords SIGNALING PROTEIN /
SIGNALING PROTEIN /  Wnt signalling
Wnt signalling Function and homology information
Function and homology information armadillo repeat domain binding / cochlea morphogenesis / head development / segment specification / WNT5A-dependent internalization of FZD4 /
armadillo repeat domain binding / cochlea morphogenesis / head development / segment specification / WNT5A-dependent internalization of FZD4 /  cell development / dorsal/ventral axis specification ...Negative regulation of TCF-dependent signaling by DVL-interacting proteins / convergent extension involved in neural plate elongation / planar cell polarity pathway involved in neural tube closure /
cell development / dorsal/ventral axis specification ...Negative regulation of TCF-dependent signaling by DVL-interacting proteins / convergent extension involved in neural plate elongation / planar cell polarity pathway involved in neural tube closure /  armadillo repeat domain binding / cochlea morphogenesis / head development / segment specification / WNT5A-dependent internalization of FZD4 /
armadillo repeat domain binding / cochlea morphogenesis / head development / segment specification / WNT5A-dependent internalization of FZD4 /  cell development / dorsal/ventral axis specification / non-canonical Wnt signaling pathway / positive regulation of neuron projection arborization / axial mesoderm formation / WNT5:FZD7-mediated leishmania damping / clathrin-coated endocytic vesicle / post-anal tail morphogenesis / APC truncation mutants have impaired AXIN binding / AXIN missense mutants destabilize the destruction complex / Truncations of AMER1 destabilize the destruction complex / epigenetic programming in the zygotic pronuclei / beta-catenin destruction complex / positive regulation of ubiquitin-dependent protein catabolic process /
cell development / dorsal/ventral axis specification / non-canonical Wnt signaling pathway / positive regulation of neuron projection arborization / axial mesoderm formation / WNT5:FZD7-mediated leishmania damping / clathrin-coated endocytic vesicle / post-anal tail morphogenesis / APC truncation mutants have impaired AXIN binding / AXIN missense mutants destabilize the destruction complex / Truncations of AMER1 destabilize the destruction complex / epigenetic programming in the zygotic pronuclei / beta-catenin destruction complex / positive regulation of ubiquitin-dependent protein catabolic process /  frizzled binding / Beta-catenin phosphorylation cascade / Signaling by GSK3beta mutants / CTNNB1 S33 mutants aren't phosphorylated / CTNNB1 S37 mutants aren't phosphorylated / CTNNB1 S45 mutants aren't phosphorylated / CTNNB1 T41 mutants aren't phosphorylated / PCP/CE pathway /
frizzled binding / Beta-catenin phosphorylation cascade / Signaling by GSK3beta mutants / CTNNB1 S33 mutants aren't phosphorylated / CTNNB1 S37 mutants aren't phosphorylated / CTNNB1 S45 mutants aren't phosphorylated / CTNNB1 T41 mutants aren't phosphorylated / PCP/CE pathway /  I-SMAD binding / Wnt signalosome / Signaling by Hippo / WNT mediated activation of DVL /
I-SMAD binding / Wnt signalosome / Signaling by Hippo / WNT mediated activation of DVL /  aggresome / Disassembly of the destruction complex and recruitment of AXIN to the membrane /
aggresome / Disassembly of the destruction complex and recruitment of AXIN to the membrane /  Wnt signaling pathway, planar cell polarity pathway / nucleocytoplasmic transport / negative regulation of protein metabolic process / negative regulation of fat cell differentiation / RUNX1 regulates transcription of genes involved in WNT signaling / RUNX1 regulates estrogen receptor mediated transcription / heart looping / outflow tract morphogenesis / negative regulation of transcription elongation by RNA polymerase II / positive regulation of transforming growth factor beta receptor signaling pathway / activation of protein kinase activity / SMAD binding /
Wnt signaling pathway, planar cell polarity pathway / nucleocytoplasmic transport / negative regulation of protein metabolic process / negative regulation of fat cell differentiation / RUNX1 regulates transcription of genes involved in WNT signaling / RUNX1 regulates estrogen receptor mediated transcription / heart looping / outflow tract morphogenesis / negative regulation of transcription elongation by RNA polymerase II / positive regulation of transforming growth factor beta receptor signaling pathway / activation of protein kinase activity / SMAD binding /  R-SMAD binding / ubiquitin-like ligase-substrate adaptor activity / lateral plasma membrane / canonical Wnt signaling pathway / signaling adaptor activity / cytoplasmic microtubule organization / positive regulation of JUN kinase activity / positive regulation of peptidyl-threonine phosphorylation / positive regulation of protein ubiquitination / Asymmetric localization of PCP proteins / TCF dependent signaling in response to WNT / neural tube closure / RHO GTPases Activate Formins / cell periphery / Degradation of DVL / regulation of actin cytoskeleton organization / Degradation of AXIN / sensory perception of sound / positive regulation of JNK cascade / Degradation of beta-catenin by the destruction complex / negative regulation of canonical Wnt signaling pathway /
R-SMAD binding / ubiquitin-like ligase-substrate adaptor activity / lateral plasma membrane / canonical Wnt signaling pathway / signaling adaptor activity / cytoplasmic microtubule organization / positive regulation of JUN kinase activity / positive regulation of peptidyl-threonine phosphorylation / positive regulation of protein ubiquitination / Asymmetric localization of PCP proteins / TCF dependent signaling in response to WNT / neural tube closure / RHO GTPases Activate Formins / cell periphery / Degradation of DVL / regulation of actin cytoskeleton organization / Degradation of AXIN / sensory perception of sound / positive regulation of JNK cascade / Degradation of beta-catenin by the destruction complex / negative regulation of canonical Wnt signaling pathway /  protein localization /
protein localization /  beta-catenin binding /
beta-catenin binding /  small GTPase binding / protein polyubiquitination / positive regulation of DNA-binding transcription factor activity / positive regulation of protein catabolic process / : / microtubule cytoskeleton / protein-macromolecule adaptor activity / positive regulation of proteasomal ubiquitin-dependent protein catabolic process /
small GTPase binding / protein polyubiquitination / positive regulation of DNA-binding transcription factor activity / positive regulation of protein catabolic process / : / microtubule cytoskeleton / protein-macromolecule adaptor activity / positive regulation of proteasomal ubiquitin-dependent protein catabolic process /  p53 binding / Cargo recognition for clathrin-mediated endocytosis / apical part of cell /
p53 binding / Cargo recognition for clathrin-mediated endocytosis / apical part of cell /  Clathrin-mediated endocytosis / positive regulation of peptidyl-serine phosphorylation / regulation of cell population proliferation /
Clathrin-mediated endocytosis / positive regulation of peptidyl-serine phosphorylation / regulation of cell population proliferation /  heart development /
heart development /  cell cortex / cytoplasmic vesicle / proteasome-mediated ubiquitin-dependent protein catabolic process / protein-containing complex assembly / Estrogen-dependent gene expression / in utero embryonic development / molecular adaptor activity /
cell cortex / cytoplasmic vesicle / proteasome-mediated ubiquitin-dependent protein catabolic process / protein-containing complex assembly / Estrogen-dependent gene expression / in utero embryonic development / molecular adaptor activity /  nuclear body / Ub-specific processing proteases / intracellular signal transduction / positive regulation of protein phosphorylation / protein domain specific binding / negative regulation of gene expression / apoptotic process
nuclear body / Ub-specific processing proteases / intracellular signal transduction / positive regulation of protein phosphorylation / protein domain specific binding / negative regulation of gene expression / apoptotic process
 Homo sapiens (human)
Homo sapiens (human) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 3.09 Å
MOLECULAR REPLACEMENT / Resolution: 3.09 Å  Authors
Authors Japan, 2items
Japan, 2items  Citation
Citation Journal: Sci.Signal. / Year: 2019
Journal: Sci.Signal. / Year: 2019 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 6jck.cif.gz
6jck.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb6jck.ent.gz
pdb6jck.ent.gz PDB format
PDB format 6jck.json.gz
6jck.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/jc/6jck
https://data.pdbj.org/pub/pdb/validation_reports/jc/6jck ftp://data.pdbj.org/pub/pdb/validation_reports/jc/6jck
ftp://data.pdbj.org/pub/pdb/validation_reports/jc/6jck Links
Links Assembly
Assembly
 Components
Components AXIN1 / Axis inhibition protein 1 / hAxin
AXIN1 / Axis inhibition protein 1 / hAxin
 Homo sapiens (human) / Gene: AXIN1, AXIN / Plasmid: pCold-GST / Production host:
Homo sapiens (human) / Gene: AXIN1, AXIN / Plasmid: pCold-GST / Production host: 
 Escherichia coli (E. coli) / References: UniProt: O15169
Escherichia coli (E. coli) / References: UniProt: O15169
 Homo sapiens (human) / Gene: DVL2 / Plasmid: pCold-GST / Production host:
Homo sapiens (human) / Gene: DVL2 / Plasmid: pCold-GST / Production host: 
 Escherichia coli (E. coli) / References: UniProt: O14641
Escherichia coli (E. coli) / References: UniProt: O14641 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation
 SYNCHROTRON / Site:
SYNCHROTRON / Site:  SPring-8
SPring-8  / Beamline: BL44XU / Wavelength: 0.9 Å
/ Beamline: BL44XU / Wavelength: 0.9 Å : 0.9 Å / Relative weight: 1
: 0.9 Å / Relative weight: 1  Processing
Processing :
:  MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller













 PDBj
PDBj










