+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6hym | ||||||
|---|---|---|---|---|---|---|---|
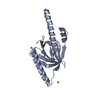
| Title | Structure of PCM1 LIR motif bound to GABARAP | ||||||
 Components Components | Pericentriolar material 1 protein,Gamma-aminobutyric acid receptor-associated protein | ||||||
 Keywords Keywords |  SIGNALING PROTEIN / SIGNALING PROTEIN /  Autophagy / Autophagy /  ATG8 / LIR ATG8 / LIR | ||||||
| Function / homology |  Function and homology information Function and homology informationprotein-containing complex localization to centriolar satellite / intraciliary transport involved in cilium assembly / interkinetic nuclear migration / microtubule anchoring / microtubule anchoring at centrosome / ciliary transition zone / positive regulation of protein K48-linked ubiquitination / regulation of Rac protein signal transduction / regulation of protein complex stability / neuronal stem cell population maintenance ...protein-containing complex localization to centriolar satellite / intraciliary transport involved in cilium assembly / interkinetic nuclear migration / microtubule anchoring / microtubule anchoring at centrosome / ciliary transition zone / positive regulation of protein K48-linked ubiquitination / regulation of Rac protein signal transduction / regulation of protein complex stability / neuronal stem cell population maintenance /  GABA receptor binding / positive regulation of intracellular protein transport / cellular response to nitrogen starvation / GABA receptor binding / positive regulation of intracellular protein transport / cellular response to nitrogen starvation /  phosphatidylethanolamine binding / non-motile cilium assembly / TBC/RABGAPs / phosphatidylethanolamine binding / non-motile cilium assembly / TBC/RABGAPs /  centrosome cycle / protein localization to centrosome / microtubule associated complex / centrosome cycle / protein localization to centrosome / microtubule associated complex /  Macroautophagy / Macroautophagy /  beta-tubulin binding / beta-tubulin binding /  pericentriolar material / pericentriolar material /  smooth endoplasmic reticulum / smooth endoplasmic reticulum /  axoneme / autophagosome membrane / axoneme / autophagosome membrane /  social behavior / social behavior /  autophagosome assembly / centriolar satellite / autophagosome assembly / centriolar satellite /  cilium assembly / extrinsic apoptotic signaling pathway via death domain receptors / cilium assembly / extrinsic apoptotic signaling pathway via death domain receptors /  protein targeting / protein targeting /  autophagosome / cytoplasmic microtubule organization / Loss of Nlp from mitotic centrosomes / Loss of proteins required for interphase microtubule organization from the centrosome / Recruitment of mitotic centrosome proteins and complexes / sperm midpiece / Recruitment of NuMA to mitotic centrosomes / Anchoring of the basal body to the plasma membrane / autophagosome / cytoplasmic microtubule organization / Loss of Nlp from mitotic centrosomes / Loss of proteins required for interphase microtubule organization from the centrosome / Recruitment of mitotic centrosome proteins and complexes / sperm midpiece / Recruitment of NuMA to mitotic centrosomes / Anchoring of the basal body to the plasma membrane /  centriole / AURKA Activation by TPX2 / ciliary basal body / centriole / AURKA Activation by TPX2 / ciliary basal body /  macroautophagy / macroautophagy /  neuron migration / microtubule cytoskeleton organization / negative regulation of neurogenesis / neuron migration / microtubule cytoskeleton organization / negative regulation of neurogenesis /  Regulation of PLK1 Activity at G2/M Transition / Regulation of PLK1 Activity at G2/M Transition /  actin cytoskeleton / positive regulation of proteasomal ubiquitin-dependent protein catabolic process / actin cytoskeleton / positive regulation of proteasomal ubiquitin-dependent protein catabolic process /  protein transport / apical part of cell / cytoplasmic vesicle / protein transport / apical part of cell / cytoplasmic vesicle /  microtubule binding / chemical synaptic transmission / microtubule binding / chemical synaptic transmission /  nuclear membrane / nuclear membrane /  microtubule / microtubule /  lysosome / molecular adaptor activity / lysosome / molecular adaptor activity /  Golgi membrane / Golgi membrane /  centrosome / centrosome /  synapse / synapse /  ubiquitin protein ligase binding / protein-containing complex / ubiquitin protein ligase binding / protein-containing complex /  nucleoplasm / nucleoplasm /  membrane / identical protein binding / membrane / identical protein binding /  plasma membrane / plasma membrane /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function | ||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.86 Å MOLECULAR REPLACEMENT / Resolution: 1.86 Å | ||||||
 Authors Authors | Mouilleron, S. / Wirth, M. / Zhang, W. / O'Reilly, N. / Tooze, S. / Johansen, T. / Razi, M. / Nyoni, L. / Joshi, D. | ||||||
 Citation Citation |  Journal: Nat Commun / Year: 2019 Journal: Nat Commun / Year: 2019Title: Molecular determinants regulating selective binding of autophagy adapters and receptors to ATG8 proteins. Authors: Wirth, M. / Zhang, W. / Razi, M. / Nyoni, L. / Joshi, D. / O'Reilly, N. / Johansen, T. / Tooze, S.A. / Mouilleron, S. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6hym.cif.gz 6hym.cif.gz | 126 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6hym.ent.gz pdb6hym.ent.gz | 98.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6hym.json.gz 6hym.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/hy/6hym https://data.pdbj.org/pub/pdb/validation_reports/hy/6hym ftp://data.pdbj.org/pub/pdb/validation_reports/hy/6hym ftp://data.pdbj.org/pub/pdb/validation_reports/hy/6hym | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  6hylC  6hynC  6hyoC  1gnuS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein |  / hPCM-1 / GABA(A) receptor-associated protein / MM46 / hPCM-1 / GABA(A) receptor-associated protein / MM46Mass: 16189.533 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: PCM1, GABARAP, FLC3B, HT004 / Production host: Homo sapiens (human) / Gene: PCM1, GABARAP, FLC3B, HT004 / Production host:   Escherichia coli (E. coli) / References: UniProt: Q15154, UniProt: O95166 Escherichia coli (E. coli) / References: UniProt: Q15154, UniProt: O95166#2: Chemical | ChemComp-GOL /  Glycerol Glycerol#3: Chemical | ChemComp-EDO / |  Ethylene glycol Ethylene glycol#4: Water | ChemComp-HOH / |  Water Water |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.79 Å3/Da / Density % sol: 55.87 % |
|---|---|
Crystal grow | Temperature: 293 K / Method: vapor diffusion, sitting drop / Details: 25% w/v PEG 1500, 0.1 M SPG pH 8.5 |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Diamond Diamond  / Beamline: I04 / Wavelength: 0.9282 Å / Beamline: I04 / Wavelength: 0.9282 Å |
| Detector | Type: DECTRIS PILATUS3 S 6M / Detector: PIXEL / Date: Jan 16, 2017 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 0.9282 Å / Relative weight: 1 : 0.9282 Å / Relative weight: 1 |
| Reflection | Resolution: 1.86→45.59 Å / Num. obs: 30131 / % possible obs: 99.5 % / Redundancy: 4.7 % / Biso Wilson estimate: 41.5 Å2 / CC1/2: 1 / Rmerge(I) obs: 0.02 / Rpim(I) all: 0.01 / Rrim(I) all: 0.02 / Net I/σ(I): 19.7 |
| Reflection shell | Resolution: 1.86→1.92 Å / Redundancy: 4.3 % / Rmerge(I) obs: 0.78 / Mean I/σ(I) obs: 1.3 / Num. unique obs: 3045 / CC1/2: 0.65 / Rpim(I) all: 0.42 / Rrim(I) all: 0.89 / % possible all: 97.7 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 1GNU Resolution: 1.86→45.587 Å / SU ML: 0.32 / Cross valid method: FREE R-VALUE / σ(F): 1.34 / Phase error: 25.22
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.86→45.587 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller










 PDBj
PDBj







