+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6dce | ||||||
|---|---|---|---|---|---|---|---|
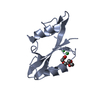
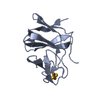
| Title | X-ray structure of FIP200 claw domain | ||||||
 Components Components | RB1-inducible coiled-coil protein 1 | ||||||
 Keywords Keywords |  STRUCTURAL PROTEIN / STRUCTURAL PROTEIN /  autophagy / autophagy /  FIP200 FIP200 | ||||||
| Function / homology |  Function and homology information Function and homology information regulation of protein lipidation / ribophagy / glycophagy / Atg1/ULK1 kinase complex / phagophore assembly site membrane / piecemeal microautophagy of the nucleus / phagophore assembly site / reticulophagy / regulation of protein lipidation / ribophagy / glycophagy / Atg1/ULK1 kinase complex / phagophore assembly site membrane / piecemeal microautophagy of the nucleus / phagophore assembly site / reticulophagy /  Macroautophagy / autophagosome membrane ... Macroautophagy / autophagosome membrane ... regulation of protein lipidation / ribophagy / glycophagy / Atg1/ULK1 kinase complex / phagophore assembly site membrane / piecemeal microautophagy of the nucleus / phagophore assembly site / reticulophagy / regulation of protein lipidation / ribophagy / glycophagy / Atg1/ULK1 kinase complex / phagophore assembly site membrane / piecemeal microautophagy of the nucleus / phagophore assembly site / reticulophagy /  Macroautophagy / autophagosome membrane / Macroautophagy / autophagosome membrane /  autophagosome assembly / positive regulation of cell size / positive regulation of autophagy / extrinsic apoptotic signaling pathway / protein-membrane adaptor activity / liver development / negative regulation of extrinsic apoptotic signaling pathway / positive regulation of JNK cascade / autophagosome assembly / positive regulation of cell size / positive regulation of autophagy / extrinsic apoptotic signaling pathway / protein-membrane adaptor activity / liver development / negative regulation of extrinsic apoptotic signaling pathway / positive regulation of JNK cascade /  autophagy / autophagy /  heart development / heart development /  nuclear membrane / nuclear membrane /  lysosome / molecular adaptor activity / positive regulation of protein phosphorylation / lysosome / molecular adaptor activity / positive regulation of protein phosphorylation /  cell cycle / negative regulation of cell population proliferation / endoplasmic reticulum membrane / cell cycle / negative regulation of cell population proliferation / endoplasmic reticulum membrane /  protein kinase binding / protein kinase binding /  cytosol cytosolSimilarity search - Function | ||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  SAD / Resolution: 1.56 Å SAD / Resolution: 1.56 Å | ||||||
 Authors Authors | Su, M.-Y. / Hurley, J.H. | ||||||
 Citation Citation |  Journal: Mol. Cell / Year: 2019 Journal: Mol. Cell / Year: 2019Title: FIP200 Claw Domain Binding to p62 Promotes Autophagosome Formation at Ubiquitin Condensates. Authors: Turco, E. / Witt, M. / Abert, C. / Bock-Bierbaum, T. / Su, M.Y. / Trapannone, R. / Sztacho, M. / Danieli, A. / Shi, X. / Zaffagnini, G. / Gamper, A. / Schuschnig, M. / Fracchiolla, D. / ...Authors: Turco, E. / Witt, M. / Abert, C. / Bock-Bierbaum, T. / Su, M.Y. / Trapannone, R. / Sztacho, M. / Danieli, A. / Shi, X. / Zaffagnini, G. / Gamper, A. / Schuschnig, M. / Fracchiolla, D. / Bernklau, D. / Romanov, J. / Hartl, M. / Hurley, J.H. / Daumke, O. / Martens, S. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6dce.cif.gz 6dce.cif.gz | 31.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6dce.ent.gz pdb6dce.ent.gz | 22.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6dce.json.gz 6dce.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/dc/6dce https://data.pdbj.org/pub/pdb/validation_reports/dc/6dce ftp://data.pdbj.org/pub/pdb/validation_reports/dc/6dce ftp://data.pdbj.org/pub/pdb/validation_reports/dc/6dce | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 11617.370 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: RB1CC1, KIAA0203, RBICC / Production host: Homo sapiens (human) / Gene: RB1CC1, KIAA0203, RBICC / Production host:   Escherichia coli (E. coli) / Strain (production host): BL21(DE3) / References: UniProt: Q8TDY2 Escherichia coli (E. coli) / Strain (production host): BL21(DE3) / References: UniProt: Q8TDY2 | ||
|---|---|---|---|
| #2: Chemical |  Sulfate Sulfate#3: Water | ChemComp-HOH / |  Water Water |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.38 Å3/Da / Density % sol: 48.24 % |
|---|---|
Crystal grow | Temperature: 292 K / Method: vapor diffusion, sitting drop / Details: 0.1 M HEPES pH 7.4, 2 M Ammonium sulfate |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ALS ALS  / Beamline: 8.3.1 / Wavelength: 0.979 Å / Beamline: 8.3.1 / Wavelength: 0.979 Å |
| Detector | Type: DECTRIS PILATUS3 S 6M / Detector: PIXEL / Date: Mar 29, 2018 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 0.979 Å / Relative weight: 1 : 0.979 Å / Relative weight: 1 |
| Reflection | Resolution: 1.56→44.63 Å / Num. obs: 16114 / % possible obs: 99.1 % / Redundancy: 2 % / Net I/σ(I): 22.1 |
| Reflection shell | Resolution: 1.56→1.62 Å |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  SAD / Resolution: 1.56→19.672 Å / SU ML: 0.14 / Cross valid method: THROUGHOUT / σ(F): 0.05 / Phase error: 22.94 SAD / Resolution: 1.56→19.672 Å / SU ML: 0.14 / Cross valid method: THROUGHOUT / σ(F): 0.05 / Phase error: 22.94
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.56→19.672 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj



