+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5yf4 | ||||||
|---|---|---|---|---|---|---|---|
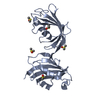
| Title | A kinase complex MST4-MOB4 | ||||||
 Components Components |
| ||||||
 Keywords Keywords |  PROTEIN BINDING / PROTEIN BINDING /  Kinase / Kinase /  Complex / Complex /  Phosphorylation Phosphorylation | ||||||
| Function / homology |  Function and homology information Function and homology information microvillus assembly / vesicle membrane / Golgi-associated vesicle / Golgi cisterna membrane / Apoptotic cleavage of cellular proteins / cellular response to starvation / negative regulation of cell migration / cell periphery / microvillus assembly / vesicle membrane / Golgi-associated vesicle / Golgi cisterna membrane / Apoptotic cleavage of cellular proteins / cellular response to starvation / negative regulation of cell migration / cell periphery /  kinase binding / cellular response to oxidative stress ... kinase binding / cellular response to oxidative stress ... microvillus assembly / vesicle membrane / Golgi-associated vesicle / Golgi cisterna membrane / Apoptotic cleavage of cellular proteins / cellular response to starvation / negative regulation of cell migration / cell periphery / microvillus assembly / vesicle membrane / Golgi-associated vesicle / Golgi cisterna membrane / Apoptotic cleavage of cellular proteins / cellular response to starvation / negative regulation of cell migration / cell periphery /  kinase binding / cellular response to oxidative stress / regulation of apoptotic process / kinase binding / cellular response to oxidative stress / regulation of apoptotic process /  dendritic spine / protein autophosphorylation / dendritic spine / protein autophosphorylation /  non-specific serine/threonine protein kinase / non-specific serine/threonine protein kinase /  protein kinase activity / intracellular signal transduction / apical plasma membrane / protein kinase activity / intracellular signal transduction / apical plasma membrane /  protein phosphorylation / protein serine kinase activity / protein serine/threonine kinase activity / neuronal cell body / apoptotic process / perinuclear region of cytoplasm / protein phosphorylation / protein serine kinase activity / protein serine/threonine kinase activity / neuronal cell body / apoptotic process / perinuclear region of cytoplasm /  Golgi apparatus / magnesium ion binding / protein homodimerization activity / extracellular exosome / Golgi apparatus / magnesium ion binding / protein homodimerization activity / extracellular exosome /  ATP binding / ATP binding /  membrane / identical protein binding / membrane / identical protein binding /  metal ion binding / metal ion binding /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function | ||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  SAD / Resolution: 1.897 Å SAD / Resolution: 1.897 Å | ||||||
 Authors Authors | Chen, M. / Zhou, Z.C. | ||||||
 Citation Citation |  Journal: J. Biol. Chem. / Year: 2018 Journal: J. Biol. Chem. / Year: 2018Title: The MST4-MOB4 complex disrupts the MST1-MOB1 complex in the Hippo-YAP pathway and plays a pro-oncogenic role in pancreatic cancer. Authors: Chen, M. / Zhang, H. / Shi, Z. / Li, Y. / Zhang, X. / Gao, Z. / Zhou, L. / Ma, J. / Xu, Q. / Guan, J. / Cheng, Y. / Jiao, S. / Zhou, Z.C. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5yf4.cif.gz 5yf4.cif.gz | 72.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5yf4.ent.gz pdb5yf4.ent.gz | 57 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5yf4.json.gz 5yf4.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/yf/5yf4 https://data.pdbj.org/pub/pdb/validation_reports/yf/5yf4 ftp://data.pdbj.org/pub/pdb/validation_reports/yf/5yf4 ftp://data.pdbj.org/pub/pdb/validation_reports/yf/5yf4 | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 18938.725 Da / Num. of mol.: 1 / Fragment: UNP residues 53-210 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: MOB4, MOB3, MOBKL3, PHOCN, PREI3, CGI-95 / Production host: Homo sapiens (human) / Gene: MOB4, MOB3, MOBKL3, PHOCN, PREI3, CGI-95 / Production host:   Escherichia coli BL21(DE3) (bacteria) / Strain (production host): BL21(DE3) / Variant (production host): CodonPlus / References: UniProt: Q9Y3A3 Escherichia coli BL21(DE3) (bacteria) / Strain (production host): BL21(DE3) / Variant (production host): CodonPlus / References: UniProt: Q9Y3A3 | ||
|---|---|---|---|
| #2: Protein/peptide |  / MST3 and SOK1-related kinase / Mammalian STE20-like protein kinase 4 / STE20-like kinase MST4 / ...MST3 and SOK1-related kinase / Mammalian STE20-like protein kinase 4 / STE20-like kinase MST4 / Serine/threonine-protein kinase MASK / MST3 and SOK1-related kinase / Mammalian STE20-like protein kinase 4 / STE20-like kinase MST4 / ...MST3 and SOK1-related kinase / Mammalian STE20-like protein kinase 4 / STE20-like kinase MST4 / Serine/threonine-protein kinase MASKMass: 2090.125 Da / Num. of mol.: 1 / Source method: obtained synthetically / Source: (synth.)   Homo sapiens (human) Homo sapiens (human)References: UniProt: Q9P289,  non-specific serine/threonine protein kinase non-specific serine/threonine protein kinase | ||
| #3: Chemical | | #4: Water | ChemComp-HOH / |  Water Water |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 1.84 Å3/Da / Density % sol: 33.03 % |
|---|---|
Crystal grow | Temperature: 289 K / Method: vapor diffusion, sitting drop / Details: 0.1M HEPES, pH 7.5, 30% PEG 1000 |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SSRF SSRF  / Beamline: BL19U1 / Wavelength: 0.97853 Å / Beamline: BL19U1 / Wavelength: 0.97853 Å |
| Detector | Type: DECTRIS PILATUS3 6M / Detector: PIXEL / Date: Dec 27, 2015 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 0.97853 Å / Relative weight: 1 : 0.97853 Å / Relative weight: 1 |
| Reflection | Resolution: 1.897→50 Å / Num. obs: 12086 / % possible obs: 98.9 % / Redundancy: 6.4 % / Rmerge(I) obs: 0.109 / Rpim(I) all: 0.046 / Rrim(I) all: 0.119 / Net I/σ(I): 15.5 |
| Reflection shell | Resolution: 1.897→1.965 Å / Redundancy: 5.5 % / Rmerge(I) obs: 0.798 / Num. unique obs: 1123 / CC1/2: 0.715 / Rpim(I) all: 0.365 / Rrim(I) all: 0.881 / % possible all: 99 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  SAD / Resolution: 1.897→30.038 Å / SU ML: 0.18 / Cross valid method: FREE R-VALUE / σ(F): 1.34 / Phase error: 19.71 / Stereochemistry target values: ML SAD / Resolution: 1.897→30.038 Å / SU ML: 0.18 / Cross valid method: FREE R-VALUE / σ(F): 1.34 / Phase error: 19.71 / Stereochemistry target values: MLDetails: The number of reflections containing anomalous reflections used in refinement is 18533. Non-anomalous reflections in data collection and refinement are 12086 and 12079 respectively.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.897→30.038 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj



