Entry Database : PDB / ID : 5xwvTitle Substrate-bound Structure of a Ketoreductase from the Second Module of the amphotericin Polyketide Synthases AmphB Keywords / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Streptomyces nodosus (bacteria)Method / / / Resolution : 1.8 Å Authors Liu, C. / Zheng, J. Funding support Organization Grant number Country National Program on Key Basic Research Project 2013CB734002 National Natural Science Foundation of China 31370101 National Natural Science Foundation of China 31570056
Journal : J. Struct. Biol. / Year : 2018Title : Substrate-bound structures of a ketoreductase from amphotericin modular polyketide synthase.Authors : Liu, C. / Yuan, M. / Xu, X. / Wang, L. / Keatinge-Clay, A.T. / Deng, Z. / Lin, S. / Zheng, J. History Deposition Jun 30, 2017 Deposition site / Processing site Revision 1.0 Jun 6, 2018 Provider / Type Revision 1.1 Jul 4, 2018 Group / Database references / Category Item / _citation.page_first / _citation.page_lastRevision 1.2 Nov 22, 2023 Group / Database references / Refinement descriptionCategory chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model / struct_ncs_dom_lim Item _database_2.pdbx_DOI / _database_2.pdbx_database_accession ... _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _struct_ncs_dom_lim.beg_auth_comp_id / _struct_ncs_dom_lim.beg_label_asym_id / _struct_ncs_dom_lim.beg_label_comp_id / _struct_ncs_dom_lim.beg_label_seq_id / _struct_ncs_dom_lim.end_auth_comp_id / _struct_ncs_dom_lim.end_label_asym_id / _struct_ncs_dom_lim.end_label_comp_id / _struct_ncs_dom_lim.end_label_seq_id
Show all Show less
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords OXIDOREDUCTASE / modular polyketide synthease / ketoreductase
OXIDOREDUCTASE / modular polyketide synthease / ketoreductase Function and homology information
Function and homology information phosphopantetheine binding / 3-oxoacyl-[acyl-carrier-protein] synthase activity / fatty acid biosynthetic process /
phosphopantetheine binding / 3-oxoacyl-[acyl-carrier-protein] synthase activity / fatty acid biosynthetic process /  nucleotide binding / identical protein binding
nucleotide binding / identical protein binding
 Streptomyces nodosus (bacteria)
Streptomyces nodosus (bacteria) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.8 Å
MOLECULAR REPLACEMENT / Resolution: 1.8 Å  Authors
Authors China, 3items
China, 3items  Citation
Citation Journal: J. Struct. Biol. / Year: 2018
Journal: J. Struct. Biol. / Year: 2018 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 5xwv.cif.gz
5xwv.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb5xwv.ent.gz
pdb5xwv.ent.gz PDB format
PDB format 5xwv.json.gz
5xwv.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/xw/5xwv
https://data.pdbj.org/pub/pdb/validation_reports/xw/5xwv ftp://data.pdbj.org/pub/pdb/validation_reports/xw/5xwv
ftp://data.pdbj.org/pub/pdb/validation_reports/xw/5xwv

 Links
Links Assembly
Assembly
 Components
Components
 Streptomyces nodosus (bacteria) / Gene: amphB / Plasmid: pET28a / Production host:
Streptomyces nodosus (bacteria) / Gene: amphB / Plasmid: pET28a / Production host: 
 Escherichia coli (E. coli) / References: UniProt: Q93NW7
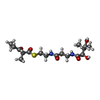
Escherichia coli (E. coli) / References: UniProt: Q93NW7 Nicotinamide adenine dinucleotide phosphate
Nicotinamide adenine dinucleotide phosphate Water
Water X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation
 SYNCHROTRON / Site:
SYNCHROTRON / Site:  SSRF
SSRF  / Beamline: BL19U1 / Wavelength: 0.97853 Å
/ Beamline: BL19U1 / Wavelength: 0.97853 Å : 0.97853 Å / Relative weight: 1
: 0.97853 Å / Relative weight: 1  Processing
Processing :
:  MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller












 PDBj
PDBj