[English] 日本語
 Yorodumi
Yorodumi- PDB-5mql: Crystal structure of dCK mutant C3S in complex with masitinib and UDP -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5mql | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of dCK mutant C3S in complex with masitinib and UDP | ||||||
 Components Components | Deoxycytidine kinase | ||||||
 Keywords Keywords |  TRANSFERASE / TRANSFERASE /  Deoxycytidine kinase / dCK / Deoxycytidine kinase / dCK /  masitinib masitinib | ||||||
| Function / homology |  Function and homology information Function and homology information deoxycytidine kinase / 2'-deoxyadenosine kinase / deoxycytidine kinase / 2'-deoxyadenosine kinase /  deoxyguanosine kinase / dAMP salvage / deoxyguanosine kinase / dAMP salvage /  deoxycytidine kinase activity / nucleoside phosphate biosynthetic process / deoxycytidine kinase activity / nucleoside phosphate biosynthetic process /  deoxyguanosine kinase activity / deoxyguanosine kinase activity /  deoxyadenosine kinase activity / Pyrimidine salvage / cytidine kinase activity ... deoxyadenosine kinase activity / Pyrimidine salvage / cytidine kinase activity ... deoxycytidine kinase / 2'-deoxyadenosine kinase / deoxycytidine kinase / 2'-deoxyadenosine kinase /  deoxyguanosine kinase / dAMP salvage / deoxyguanosine kinase / dAMP salvage /  deoxycytidine kinase activity / nucleoside phosphate biosynthetic process / deoxycytidine kinase activity / nucleoside phosphate biosynthetic process /  deoxyguanosine kinase activity / deoxyguanosine kinase activity /  deoxyadenosine kinase activity / Pyrimidine salvage / cytidine kinase activity / pyrimidine nucleotide metabolic process / Purine salvage / deoxyadenosine kinase activity / Pyrimidine salvage / cytidine kinase activity / pyrimidine nucleotide metabolic process / Purine salvage /  phosphorylation / protein homodimerization activity / phosphorylation / protein homodimerization activity /  mitochondrion / mitochondrion /  nucleoplasm / nucleoplasm /  ATP binding / ATP binding /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function | ||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 3.25 Å MOLECULAR REPLACEMENT / Resolution: 3.25 Å | ||||||
 Authors Authors | Rebuffet, E. / Hammam, K. / Saez-Ayala, M. / Gros, L. / Lopez, S. / Hajem, B. / Humbert, M. / Baudelet, E. / Audebert, S. / Betzi, S. ...Rebuffet, E. / Hammam, K. / Saez-Ayala, M. / Gros, L. / Lopez, S. / Hajem, B. / Humbert, M. / Baudelet, E. / Audebert, S. / Betzi, S. / Lugari, A. / Combes, S. / Pez, D. / Letard, S. / Mansfield, C. / Moussy, A. / de Sepulveda, P. / Morelli, X. / Dubreuil, P. | ||||||
| Funding support |  France, 1items France, 1items
| ||||||
 Citation Citation |  Journal: Nat Commun / Year: 2017 Journal: Nat Commun / Year: 2017Title: Dual protein kinase and nucleoside kinase modulators for rationally designed polypharmacology. Authors: Hammam, K. / Saez-Ayala, M. / Rebuffet, E. / Gros, L. / Lopez, S. / Hajem, B. / Humbert, M. / Baudelet, E. / Audebert, S. / Betzi, S. / Lugari, A. / Combes, S. / Letard, S. / Casteran, N. / ...Authors: Hammam, K. / Saez-Ayala, M. / Rebuffet, E. / Gros, L. / Lopez, S. / Hajem, B. / Humbert, M. / Baudelet, E. / Audebert, S. / Betzi, S. / Lugari, A. / Combes, S. / Letard, S. / Casteran, N. / Mansfield, C. / Moussy, A. / De Sepulveda, P. / Morelli, X. / Dubreuil, P. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5mql.cif.gz 5mql.cif.gz | 395.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5mql.ent.gz pdb5mql.ent.gz | 324 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5mql.json.gz 5mql.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/mq/5mql https://data.pdbj.org/pub/pdb/validation_reports/mq/5mql ftp://data.pdbj.org/pub/pdb/validation_reports/mq/5mql ftp://data.pdbj.org/pub/pdb/validation_reports/mq/5mql | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  5mqjC  5mqtC  1p60S S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 4 molecules ABCD
| #1: Protein |  / dCK / dCKMass: 34023.195 Da / Num. of mol.: 4 / Mutation: C9S,C45S,C59S Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: DCK / Production host: Homo sapiens (human) / Gene: DCK / Production host:   Escherichia coli (E. coli) / References: UniProt: P27707, Escherichia coli (E. coli) / References: UniProt: P27707,  deoxycytidine kinase deoxycytidine kinase |
|---|
-Non-polymers , 5 types, 26 molecules 
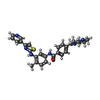
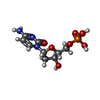






| #2: Chemical | ChemComp-UDP /  Uridine diphosphate Uridine diphosphate#3: Chemical | ChemComp-G65 / |  Masitinib Masitinib#4: Chemical |  Deoxycytidine monophosphate Deoxycytidine monophosphate#5: Chemical | ChemComp-MG / | #6: Water | ChemComp-HOH / |  Water Water |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.45 Å3/Da / Density % sol: 49.87 % |
|---|---|
Crystal grow | Temperature: 291.15 K / Method: vapor diffusion, hanging drop / pH: 5.5 Details: 100 mM sodium acetate, 20% PEG 3350, 50 mM NaCl, 24h soaking with 10 mM Masitinib |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: ID23-1 / Wavelength: 0.885602 Å / Beamline: ID23-1 / Wavelength: 0.885602 Å |
| Detector | Type: DECTRIS PILATUS3 6M / Detector: PIXEL / Date: Apr 26, 2015 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 0.885602 Å / Relative weight: 1 : 0.885602 Å / Relative weight: 1 |
| Reflection | Resolution: 3.25→42.8 Å / Num. obs: 22378 / % possible obs: 100 % / Redundancy: 8.2 % / Biso Wilson estimate: 138.18 Å2 / CC1/2: 0.994 / Rmerge(I) obs: 0.2677 / Net I/σ(I): 11.94 |
| Reflection shell | Resolution: 3.25→3.366 Å / Redundancy: 8.6 % / Num. unique obs: 2164 / CC1/2: 0.832 / % possible all: 100 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 1P60 Resolution: 3.25→47.8 Å / Cor.coef. Fo:Fc: 0.887 / Cor.coef. Fo:Fc free: 0.858 / Cross valid method: THROUGHOUT / σ(F): 0 / SU Rfree Blow DPI: 0.519
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 97.83 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.47 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: 1 / Resolution: 3.25→47.8 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 3.25→3.41 Å / Total num. of bins used: 11
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj







