[English] 日本語
 Yorodumi
Yorodumi- PDB-5m0e: Structure-based evolution of a hybrid steroid series of Autotaxin... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5m0e | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure-based evolution of a hybrid steroid series of Autotaxin inhibitors | |||||||||
 Components Components | Ectonucleotide pyrophosphatase/phosphodiesterase family member 2 | |||||||||
 Keywords Keywords |  HYDROLASE / HYDROLASE /  medicinal chemistry / structure based design / medicinal chemistry / structure based design /  autotaxin autotaxin | |||||||||
| Function / homology |  Function and homology information Function and homology informationresponse to polycyclic arene /  alkylglycerophosphoethanolamine phosphodiesterase / sphingolipid catabolic process / phospholipid catabolic process / phosphatidylcholine catabolic process / alkylglycerophosphoethanolamine phosphodiesterase / sphingolipid catabolic process / phospholipid catabolic process / phosphatidylcholine catabolic process /  lysophospholipase activity / positive regulation of lamellipodium morphogenesis / lysophospholipase activity / positive regulation of lamellipodium morphogenesis /  phosphodiesterase I activity / scavenger receptor activity / phosphodiesterase I activity / scavenger receptor activity /  alkylglycerophosphoethanolamine phosphodiesterase activity ...response to polycyclic arene / alkylglycerophosphoethanolamine phosphodiesterase activity ...response to polycyclic arene /  alkylglycerophosphoethanolamine phosphodiesterase / sphingolipid catabolic process / phospholipid catabolic process / phosphatidylcholine catabolic process / alkylglycerophosphoethanolamine phosphodiesterase / sphingolipid catabolic process / phospholipid catabolic process / phosphatidylcholine catabolic process /  lysophospholipase activity / positive regulation of lamellipodium morphogenesis / lysophospholipase activity / positive regulation of lamellipodium morphogenesis /  phosphodiesterase I activity / scavenger receptor activity / phosphodiesterase I activity / scavenger receptor activity /  alkylglycerophosphoethanolamine phosphodiesterase activity / alkylglycerophosphoethanolamine phosphodiesterase activity /  polysaccharide binding / positive regulation of oligodendrocyte differentiation / negative regulation of cell-matrix adhesion / positive regulation of epithelial cell migration / positive regulation of focal adhesion assembly / polysaccharide binding / positive regulation of oligodendrocyte differentiation / negative regulation of cell-matrix adhesion / positive regulation of epithelial cell migration / positive regulation of focal adhesion assembly /  estrous cycle / phospholipid metabolic process / positive regulation of substrate adhesion-dependent cell spreading / estrous cycle / phospholipid metabolic process / positive regulation of substrate adhesion-dependent cell spreading /  regulation of cell migration / cellular response to cadmium ion / cell chemotaxis / cellular response to estradiol stimulus / positive regulation of peptidyl-tyrosine phosphorylation / regulation of cell migration / cellular response to cadmium ion / cell chemotaxis / cellular response to estradiol stimulus / positive regulation of peptidyl-tyrosine phosphorylation /  nucleic acid binding / nucleic acid binding /  immune response / immune response /  calcium ion binding / positive regulation of cell population proliferation / calcium ion binding / positive regulation of cell population proliferation /  extracellular space / zinc ion binding extracellular space / zinc ion bindingSimilarity search - Function | |||||||||
| Biological species |   Rattus norvegicus (Norway rat) Rattus norvegicus (Norway rat) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.95 Å MOLECULAR REPLACEMENT / Resolution: 1.95 Å | |||||||||
 Authors Authors | Keune, W.-J. / Heidebrecht, T. / Perrakis, A. | |||||||||
 Citation Citation |  Journal: J. Med. Chem. / Year: 2017 Journal: J. Med. Chem. / Year: 2017Title: Rational Design of Autotaxin Inhibitors by Structural Evolution of Endogenous Modulators. Authors: Keune, W.J. / Potjewyd, F. / Heidebrecht, T. / Salgado-Polo, F. / Macdonald, S.J. / Chelvarajan, L. / Abdel Latif, A. / Soman, S. / Morris, A.J. / Watson, A.J. / Jamieson, C. / Perrakis, A. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5m0e.cif.gz 5m0e.cif.gz | 337.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5m0e.ent.gz pdb5m0e.ent.gz | 269.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5m0e.json.gz 5m0e.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/m0/5m0e https://data.pdbj.org/pub/pdb/validation_reports/m0/5m0e ftp://data.pdbj.org/pub/pdb/validation_reports/m0/5m0e ftp://data.pdbj.org/pub/pdb/validation_reports/m0/5m0e | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  5m0dC  5m0mC  5m0sC  2xr9S S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Protein / Sugars , 2 types, 2 molecules A
| #1: Protein | Mass: 95037.656 Da / Num. of mol.: 1 / Mutation: N410A, L581F, R591T, N806A Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Rattus norvegicus (Norway rat) / Gene: Enpp2, Atx, Npps2 / Cell line (production host): Hek293 / Organ (production host): kidney / Production host: Rattus norvegicus (Norway rat) / Gene: Enpp2, Atx, Npps2 / Cell line (production host): Hek293 / Organ (production host): kidney / Production host:   Homo sapiens (human) Homo sapiens (human)References: UniProt: Q64610,  alkylglycerophosphoethanolamine phosphodiesterase alkylglycerophosphoethanolamine phosphodiesterase |
|---|---|
| #2: Polysaccharide | beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta- ...beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose / Mass: 586.542 Da / Num. of mol.: 1 / Mass: 586.542 Da / Num. of mol.: 1Source method: isolated from a genetically manipulated source |
-Non-polymers , 9 types, 268 molecules 



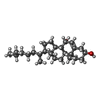
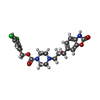











| #3: Chemical | | #4: Chemical | ChemComp-IOD /  Iodide Iodide#5: Chemical | ChemComp-CA / | #6: Chemical | #7: Chemical | ChemComp-5JK / |  7α-Hydroxycholesterol 7α-Hydroxycholesterol#8: Chemical | ChemComp-7CR / [ | #9: Chemical | ChemComp-SCN /  Thiocyanate Thiocyanate#10: Chemical | ChemComp-GOL /  Glycerol Glycerol#11: Water | ChemComp-HOH / |  Water Water |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.13 Å3/Da / Density % sol: 42.13 % |
|---|---|
Crystal grow | Temperature: 293 K / Method: vapor diffusion, hanging drop / Details: PEG 3350, NaSCN and NH4I |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SLS SLS  / Beamline: X06DA / Wavelength: 1 Å / Beamline: X06DA / Wavelength: 1 Å |
| Detector | Type: DECTRIS PILATUS 2M / Detector: PIXEL / Date: Jun 9, 2015 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 1 Å / Relative weight: 1 : 1 Å / Relative weight: 1 |
| Reflection | Resolution: 1.95→61.15 Å / Num. obs: 53155 / % possible obs: 98.8 % / Redundancy: 3.9 % / Net I/σ(I): 18.9 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 2XR9 Resolution: 1.95→61.15 Å / Cor.coef. Fo:Fc: 0.954 / Cor.coef. Fo:Fc free: 0.926 / SU B: 9.288 / SU ML: 0.127 / Cross valid method: THROUGHOUT / ESU R: 0.182 / ESU R Free: 0.158 / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 28.509 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: 1 / Resolution: 1.95→61.15 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj








