[English] 日本語
 Yorodumi
Yorodumi- PDB-5hhf: Crystal structure of Aspergillus fumigatus UDP-Galactopyranose mu... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5hhf | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Crystal structure of Aspergillus fumigatus UDP-Galactopyranose mutase mutant H63A with covalent FAD-Galactopyranose and bound UDP | |||||||||
 Components Components | UDP-galactopyranose mutase | |||||||||
 Keywords Keywords |  ISOMERASE / ISOMERASE /  NUCLEOTIDE BINDING / NUCLEOTIDE BINDING /  MUTASE / MUTASE /  FLAVIN ADENINE DINUCLEOTIDE BINDING / covalent reaction intermediate FLAVIN ADENINE DINUCLEOTIDE BINDING / covalent reaction intermediate | |||||||||
| Function / homology |  Function and homology information Function and homology information UDP-galactopyranose mutase / UDP-galactopyranose mutase /  UDP-galactopyranose mutase activity / UDP-galactopyranose mutase activity /  nucleotide binding nucleotide bindingSimilarity search - Function | |||||||||
| Biological species |   Neosartorya fumigata (mold) Neosartorya fumigata (mold) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  FOURIER SYNTHESIS / Resolution: 2.3 Å FOURIER SYNTHESIS / Resolution: 2.3 Å | |||||||||
 Authors Authors | Tanner, J.J. | |||||||||
| Funding support |  United States, 2items United States, 2items
| |||||||||
 Citation Citation |  Journal: Biochemistry / Year: 2016 Journal: Biochemistry / Year: 2016Title: In Crystallo Capture of a Covalent Intermediate in the UDP-Galactopyranose Mutase Reaction. Authors: Mehra-Chaudhary, R. / Dai, Y. / Sobrado, P. / Tanner, J.J. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5hhf.cif.gz 5hhf.cif.gz | 795 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5hhf.ent.gz pdb5hhf.ent.gz | 657.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5hhf.json.gz 5hhf.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/hh/5hhf https://data.pdbj.org/pub/pdb/validation_reports/hh/5hhf ftp://data.pdbj.org/pub/pdb/validation_reports/hh/5hhf ftp://data.pdbj.org/pub/pdb/validation_reports/hh/5hhf | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  3uthS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||||||||
| Unit cell |
| |||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS domain:
|
- Components
Components
-Protein , 1 types, 4 molecules ABCD
| #1: Protein |  Mass: 57061.422 Da / Num. of mol.: 4 / Mutation: H63A Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Neosartorya fumigata (mold) / Gene: glf, glfA / Production host: Neosartorya fumigata (mold) / Gene: glf, glfA / Production host:   Escherichia coli (E. coli) / References: UniProt: Q4W1X2, Escherichia coli (E. coli) / References: UniProt: Q4W1X2,  UDP-galactopyranose mutase UDP-galactopyranose mutase |
|---|
-Non-polymers , 6 types, 760 molecules 
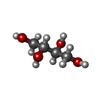









| #2: Chemical | ChemComp-SO4 /  Sulfate Sulfate#3: Chemical | ChemComp-MRY /  Erythritol Erythritol#4: Chemical | #5: Chemical |  Uridine diphosphate Uridine diphosphate#6: Chemical | #7: Water | ChemComp-HOH / |  Water Water |
|---|
-Details
| Sequence details | AUTHOR STATES THAT A429T IS AN UNINTENDED |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 4.8 Å3/Da / Density % sol: 74.37 % |
|---|---|
Crystal grow | Temperature: 293 K / Method: vapor diffusion, sitting drop / pH: 4.5 Details: 1.6 M ammonium sulfate and 0.1 M sodium acetate at pH 4.5 |
-Data collection
| Diffraction | Mean temperature: 100 K | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ALS ALS  / Beamline: 4.2.2 / Wavelength: 1 Å / Beamline: 4.2.2 / Wavelength: 1 Å | |||||||||||||||||||||||||||
| Detector | Type: RDI CMOS_8M / Detector: CMOS / Date: Sep 4, 2015 | |||||||||||||||||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | |||||||||||||||||||||||||||
| Radiation wavelength | Wavelength : 1 Å / Relative weight: 1 : 1 Å / Relative weight: 1 | |||||||||||||||||||||||||||
| Reflection | Resolution: 2.3→61.61 Å / Num. obs: 196082 / % possible obs: 100 % / Redundancy: 21.1 % / Biso Wilson estimate: 32.33 Å2 / CC1/2: 0.998 / Rmerge(I) obs: 0.201 / Rpim(I) all: 0.045 / Net I/σ(I): 15.1 / Num. measured all: 4132787 | |||||||||||||||||||||||||||
| Reflection shell | Diffraction-ID: 1 / Rejects: 0
|
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  FOURIER SYNTHESIS FOURIER SYNTHESISStarting model: 3uth Resolution: 2.3→61.61 Å / SU ML: 0.25 / Cross valid method: FREE R-VALUE / σ(F): 1.89 / Phase error: 20.95 / Stereochemistry target values: ML
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 127.55 Å2 / Biso mean: 39.5653 Å2 / Biso min: 18.74 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 2.3→61.61 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints NCS |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Total num. of bins used: 27 / % reflection obs: 100 %
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj







