Entry Database : PDB / ID : 5cuhTitle Crystal structure MMP-9 complexes with a constrained hydroxamate based inhibitor LT4 Matrix metalloproteinase-9,Matrix metalloproteinase-9 Keywords / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Homo sapiens (human)Method / / / Resolution : 1.83 Å Authors Tepshi, L. / Vera, L. / Nuti, E. / Rosalia, L. / Rossello, A. / Stura, E.A. Journal : Eur.J.Med.Chem. / Year : 2016Title : Discovery of a new selective inhibitor of A Disintegrin And Metalloprotease 10 (ADAM-10) able to reduce the shedding of NKG2D ligands in Hodgkin's lymphoma cell models.Authors : Camodeca, C. / Nuti, E. / Tepshi, L. / Boero, S. / Tuccinardi, T. / Stura, E.A. / Poggi, A. / Zocchi, M.R. / Rossello, A. History Deposition Jul 24, 2015 Deposition site / Processing site Revision 1.0 Feb 10, 2016 Provider / Type Revision 1.1 Apr 20, 2016 Group Revision 1.2 Sep 27, 2017 Group / Database references / Derived calculationsCategory / diffrn_detector / pdbx_struct_oper_listItem / _diffrn_detector.detector / _pdbx_struct_oper_list.symmetry_operationRevision 1.3 Nov 8, 2023 Group Data collection / Database references ... Data collection / Database references / Derived calculations / Refinement description Category chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model / pdbx_struct_conn_angle / struct_conn Item _database_2.pdbx_DOI / _database_2.pdbx_database_accession ... _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _pdbx_struct_conn_angle.ptnr1_auth_seq_id / _pdbx_struct_conn_angle.ptnr3_auth_seq_id / _pdbx_struct_conn_angle.value / _struct_conn.pdbx_dist_value / _struct_conn.pdbx_ptnr1_label_alt_id / _struct_conn.ptnr1_auth_asym_id / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr2_auth_asym_id / _struct_conn.ptnr2_auth_comp_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id / _struct_conn.ptnr2_label_atom_id / _struct_conn.ptnr2_label_comp_id
Show all Show less
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords HYDROLASE / MMP-9 hydroxamate-based inhibitor gelatinase
HYDROLASE / MMP-9 hydroxamate-based inhibitor gelatinase Function and homology information
Function and homology information gelatinase B / negative regulation of epithelial cell differentiation involved in kidney development / negative regulation of cation channel activity / negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway / cellular response to UV-A / positive regulation of keratinocyte migration / regulation of neuroinflammatory response / positive regulation of epidermal growth factor receptor signaling pathway / Assembly of collagen fibrils and other multimeric structures / endodermal cell differentiation ...
gelatinase B / negative regulation of epithelial cell differentiation involved in kidney development / negative regulation of cation channel activity / negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway / cellular response to UV-A / positive regulation of keratinocyte migration / regulation of neuroinflammatory response / positive regulation of epidermal growth factor receptor signaling pathway / Assembly of collagen fibrils and other multimeric structures / endodermal cell differentiation ... gelatinase B / negative regulation of epithelial cell differentiation involved in kidney development / negative regulation of cation channel activity / negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway / cellular response to UV-A / positive regulation of keratinocyte migration / regulation of neuroinflammatory response / positive regulation of epidermal growth factor receptor signaling pathway / Assembly of collagen fibrils and other multimeric structures / endodermal cell differentiation / Activation of Matrix Metalloproteinases / positive regulation of release of cytochrome c from mitochondria / response to amyloid-beta / macrophage differentiation / Collagen degradation / collagen catabolic process / EPH-ephrin mediated repulsion of cells / extracellular matrix disassembly / ephrin receptor signaling pathway / positive regulation of DNA binding / negative regulation of intrinsic apoptotic signaling pathway / positive regulation of vascular associated smooth muscle cell proliferation / cellular response to cadmium ion /
gelatinase B / negative regulation of epithelial cell differentiation involved in kidney development / negative regulation of cation channel activity / negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway / cellular response to UV-A / positive regulation of keratinocyte migration / regulation of neuroinflammatory response / positive regulation of epidermal growth factor receptor signaling pathway / Assembly of collagen fibrils and other multimeric structures / endodermal cell differentiation / Activation of Matrix Metalloproteinases / positive regulation of release of cytochrome c from mitochondria / response to amyloid-beta / macrophage differentiation / Collagen degradation / collagen catabolic process / EPH-ephrin mediated repulsion of cells / extracellular matrix disassembly / ephrin receptor signaling pathway / positive regulation of DNA binding / negative regulation of intrinsic apoptotic signaling pathway / positive regulation of vascular associated smooth muscle cell proliferation / cellular response to cadmium ion /  collagen binding /
collagen binding /  embryo implantation / Degradation of the extracellular matrix / extracellular matrix organization / positive regulation of receptor binding /
embryo implantation / Degradation of the extracellular matrix / extracellular matrix organization / positive regulation of receptor binding /  skeletal system development / Signaling by SCF-KIT /
skeletal system development / Signaling by SCF-KIT /  metalloendopeptidase activity / cellular response to reactive oxygen species /
metalloendopeptidase activity / cellular response to reactive oxygen species /  metallopeptidase activity /
metallopeptidase activity /  cell migration / tertiary granule lumen /
cell migration / tertiary granule lumen /  peptidase activity / Interleukin-4 and Interleukin-13 signaling / collagen-containing extracellular matrix /
peptidase activity / Interleukin-4 and Interleukin-13 signaling / collagen-containing extracellular matrix /  endopeptidase activity / cellular response to lipopolysaccharide / ficolin-1-rich granule lumen / Extra-nuclear estrogen signaling / positive regulation of protein phosphorylation / positive regulation of apoptotic process / serine-type endopeptidase activity / apoptotic process / Neutrophil degranulation / negative regulation of apoptotic process /
endopeptidase activity / cellular response to lipopolysaccharide / ficolin-1-rich granule lumen / Extra-nuclear estrogen signaling / positive regulation of protein phosphorylation / positive regulation of apoptotic process / serine-type endopeptidase activity / apoptotic process / Neutrophil degranulation / negative regulation of apoptotic process /  proteolysis /
proteolysis /  extracellular space / extracellular exosome / zinc ion binding / extracellular region / identical protein binding
extracellular space / extracellular exosome / zinc ion binding / extracellular region / identical protein binding
 Homo sapiens (human)
Homo sapiens (human) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.83 Å
MOLECULAR REPLACEMENT / Resolution: 1.83 Å  Authors
Authors Citation
Citation Journal: Eur.J.Med.Chem. / Year: 2016
Journal: Eur.J.Med.Chem. / Year: 2016 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 5cuh.cif.gz
5cuh.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb5cuh.ent.gz
pdb5cuh.ent.gz PDB format
PDB format 5cuh.json.gz
5cuh.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/cu/5cuh
https://data.pdbj.org/pub/pdb/validation_reports/cu/5cuh ftp://data.pdbj.org/pub/pdb/validation_reports/cu/5cuh
ftp://data.pdbj.org/pub/pdb/validation_reports/cu/5cuh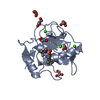
 Links
Links Assembly
Assembly


 Components
Components
 Homo sapiens (human) / Gene: MMP9, CLG4B / Plasmid: pET-14b / Production host:
Homo sapiens (human) / Gene: MMP9, CLG4B / Plasmid: pET-14b / Production host: 
 ESCHERICHIA COLI (E. coli) / Strain (production host): Bl21 (de3 Star) / References: UniProt: P14780,
ESCHERICHIA COLI (E. coli) / Strain (production host): Bl21 (de3 Star) / References: UniProt: P14780,  gelatinase B
gelatinase B











 Ethylene glycol
Ethylene glycol Propanediol
Propanediol Dimethyl sulfoxide
Dimethyl sulfoxide Water
Water X-RAY DIFFRACTION
X-RAY DIFFRACTION Sample preparation
Sample preparation
 SYNCHROTRON / Site:
SYNCHROTRON / Site:  SOLEIL
SOLEIL  / Beamline: PROXIMA 2 / Wavelength: 0.9801 Å
/ Beamline: PROXIMA 2 / Wavelength: 0.9801 Å : 0.9801 Å / Relative weight: 1
: 0.9801 Å / Relative weight: 1  Processing
Processing :
:  MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller












 PDBj
PDBj















