Entry Database : PDB / ID : 5aq0Title The structure of the Transthyretin-like domain of the first catalytic domain of the HUMAN Carboxypeptidase D CARBOXYPEPTIDASE D Keywords / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species HOMO SAPIENS (human)Method / / / Resolution : 0.95 Å Authors Gallego, P. / Garcia-Pardo, J. / Lorenzo, J. / Aviles, F.X. / Ventura, S. / Reverter, D. Journal : To be Published Title : The Structure of the Ttldomain of the Human Carboxypeptidase DAuthors : Gallego, P. / Garcia-Pardo, J. / Berenguer, E. / Lorenzo, J. / Aviles, F.X. / Ventura, S. / Reverter, D. History Deposition Sep 18, 2015 Deposition site / Processing site Revision 1.0 Sep 28, 2016 Provider / Type Revision 1.1 May 8, 2024 Group Data collection / Database references ... Data collection / Database references / Derived calculations / Other Category chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / pdbx_database_status / struct_site Item _database_2.pdbx_DOI / _database_2.pdbx_database_accession ... _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _pdbx_database_status.status_code_sf / _struct_site.pdbx_auth_asym_id / _struct_site.pdbx_auth_comp_id / _struct_site.pdbx_auth_seq_id
Show all Show less
 Yorodumi
Yorodumi Open data
Open data Basic information
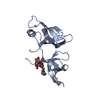
Basic information Components
Components
 Keywords
Keywords HYDROLASE / MAN CARBOXYPEPTIDASE D / TTL / TRANSTHYRETIN-LIKE DOMAIN
HYDROLASE / MAN CARBOXYPEPTIDASE D / TTL / TRANSTHYRETIN-LIKE DOMAIN Function and homology information
Function and homology information metallocarboxypeptidase D / serine-type carboxypeptidase activity / peptide metabolic process / Golgi Associated Vesicle Biogenesis /
metallocarboxypeptidase D / serine-type carboxypeptidase activity / peptide metabolic process / Golgi Associated Vesicle Biogenesis /  metallocarboxypeptidase activity / protein processing / membrane => GO:0016020 /
metallocarboxypeptidase activity / protein processing / membrane => GO:0016020 /  extracellular space / extracellular exosome / zinc ion binding ...
extracellular space / extracellular exosome / zinc ion binding ... metallocarboxypeptidase D / serine-type carboxypeptidase activity / peptide metabolic process / Golgi Associated Vesicle Biogenesis /
metallocarboxypeptidase D / serine-type carboxypeptidase activity / peptide metabolic process / Golgi Associated Vesicle Biogenesis /  metallocarboxypeptidase activity / protein processing / membrane => GO:0016020 /
metallocarboxypeptidase activity / protein processing / membrane => GO:0016020 /  extracellular space / extracellular exosome / zinc ion binding /
extracellular space / extracellular exosome / zinc ion binding /  membrane /
membrane /  plasma membrane
plasma membrane
 HOMO SAPIENS (human)
HOMO SAPIENS (human) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 0.95 Å
MOLECULAR REPLACEMENT / Resolution: 0.95 Å  Authors
Authors Citation
Citation Journal: To be Published
Journal: To be Published Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 5aq0.cif.gz
5aq0.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb5aq0.ent.gz
pdb5aq0.ent.gz PDB format
PDB format 5aq0.json.gz
5aq0.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/aq/5aq0
https://data.pdbj.org/pub/pdb/validation_reports/aq/5aq0 ftp://data.pdbj.org/pub/pdb/validation_reports/aq/5aq0
ftp://data.pdbj.org/pub/pdb/validation_reports/aq/5aq0 Links
Links Assembly
Assembly
 Components
Components / METALLOCARBOXYPEPTIDASE D / GP180 / METALLOCARBOXYPEPTIDASE D / GP180 / HUMAN CARBOXYPEPTIDASE D / ...METALLOCARBOXYPEPTIDASE D / GP180 / METALLOCARBOXYPEPTIDASE D / GP180 / HUMAN CARBOXYPEPTIDASE D / HUMAN CARBOXYPEPTIDASE D
/ METALLOCARBOXYPEPTIDASE D / GP180 / METALLOCARBOXYPEPTIDASE D / GP180 / HUMAN CARBOXYPEPTIDASE D / ...METALLOCARBOXYPEPTIDASE D / GP180 / METALLOCARBOXYPEPTIDASE D / GP180 / HUMAN CARBOXYPEPTIDASE D / HUMAN CARBOXYPEPTIDASE D
 HOMO SAPIENS (human) / Production host:
HOMO SAPIENS (human) / Production host: 
 ESCHERICHIA COLI (E. coli) / Strain (production host): BL21 / References: UniProt: O75976,
ESCHERICHIA COLI (E. coli) / Strain (production host): BL21 / References: UniProt: O75976,  metallocarboxypeptidase D
metallocarboxypeptidase D Glycerol
Glycerol Water
Water X-RAY DIFFRACTION / Number of used crystals: 3
X-RAY DIFFRACTION / Number of used crystals: 3  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  ALBA
ALBA  / Beamline: XALOC / Wavelength: 0.979491
/ Beamline: XALOC / Wavelength: 0.979491  : 0.979491 Å / Relative weight: 1
: 0.979491 Å / Relative weight: 1  Processing
Processing :
:  MOLECULAR REPLACEMENT / Resolution: 0.95→42.73 Å / Cor.coef. Fo:Fc: 0.974 / Cor.coef. Fo:Fc free: 0.968 / SU B: 0.627 / SU ML: 0.016 / Cross valid method: THROUGHOUT / ESU R: 0.022 / ESU R Free: 0.023 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS.
MOLECULAR REPLACEMENT / Resolution: 0.95→42.73 Å / Cor.coef. Fo:Fc: 0.974 / Cor.coef. Fo:Fc free: 0.968 / SU B: 0.627 / SU ML: 0.016 / Cross valid method: THROUGHOUT / ESU R: 0.022 / ESU R Free: 0.023 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS. Movie
Movie Controller
Controller











 PDBj
PDBj



