+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4xc2 | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Crystal structure of GABARAP in complex with KBTBD6 LIR peptide | |||||||||||||||||||||
 Components Components |
| |||||||||||||||||||||
 Keywords Keywords |  IMMUNE SYSTEM / IMMUNE SYSTEM /  autophagy / autophagy /  complex complex | |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of protein K48-linked ubiquitination / regulation of Rac protein signal transduction /  GABA receptor binding / cellular response to nitrogen starvation / GABA receptor binding / cellular response to nitrogen starvation /  phosphatidylethanolamine binding / TBC/RABGAPs / microtubule associated complex / phosphatidylethanolamine binding / TBC/RABGAPs / microtubule associated complex /  Macroautophagy / Macroautophagy /  beta-tubulin binding / beta-tubulin binding /  axoneme ...positive regulation of protein K48-linked ubiquitination / regulation of Rac protein signal transduction / axoneme ...positive regulation of protein K48-linked ubiquitination / regulation of Rac protein signal transduction /  GABA receptor binding / cellular response to nitrogen starvation / GABA receptor binding / cellular response to nitrogen starvation /  phosphatidylethanolamine binding / TBC/RABGAPs / microtubule associated complex / phosphatidylethanolamine binding / TBC/RABGAPs / microtubule associated complex /  Macroautophagy / Macroautophagy /  beta-tubulin binding / beta-tubulin binding /  axoneme / autophagosome membrane / axoneme / autophagosome membrane /  autophagosome assembly / extrinsic apoptotic signaling pathway via death domain receptors / autophagosome assembly / extrinsic apoptotic signaling pathway via death domain receptors /  smooth endoplasmic reticulum / smooth endoplasmic reticulum /  autophagosome / autophagosome /  protein targeting / sperm midpiece / protein targeting / sperm midpiece /  post-translational protein modification / post-translational protein modification /  macroautophagy / macroautophagy /  autophagy / microtubule cytoskeleton organization / autophagy / microtubule cytoskeleton organization /  actin cytoskeleton / positive regulation of proteasomal ubiquitin-dependent protein catabolic process / Antigen processing: Ubiquitination & Proteasome degradation / actin cytoskeleton / positive regulation of proteasomal ubiquitin-dependent protein catabolic process / Antigen processing: Ubiquitination & Proteasome degradation /  protein transport / protein transport /  Neddylation / Neddylation /  cell body / cytoplasmic vesicle / cell body / cytoplasmic vesicle /  microtubule binding / chemical synaptic transmission / microtubule binding / chemical synaptic transmission /  microtubule / microtubule /  lysosome / lysosome /  Golgi membrane / Golgi membrane /  synapse / synapse /  ubiquitin protein ligase binding / perinuclear region of cytoplasm / ubiquitin protein ligase binding / perinuclear region of cytoplasm /  Golgi apparatus / Golgi apparatus /  plasma membrane / plasma membrane /  cytosol cytosolSimilarity search - Function | |||||||||||||||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||||||||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.9 Å MOLECULAR REPLACEMENT / Resolution: 1.9 Å | |||||||||||||||||||||
 Authors Authors | Huber, J. / Genau, H.M. / Baschieri, F. / Doetsch, V. / Farhan, H. / Rogov, V.V. / Behrends, C. / Akutsu, M. | |||||||||||||||||||||
| Funding support |  Germany, 6items Germany, 6items
| |||||||||||||||||||||
 Citation Citation |  Journal: Mol.Cell / Year: 2015 Journal: Mol.Cell / Year: 2015Title: CUL3-KBTBD6/KBTBD7 Ubiquitin Ligase Cooperates with GABARAP Proteins to Spatially Restrict TIAM1-RAC1 Signaling. Authors: Genau, H.M. / Huber, J. / Baschieri, F. / Akutsu, M. / Dotsch, V. / Farhan, H. / Rogov, V. / Behrends, C. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4xc2.cif.gz 4xc2.cif.gz | 118.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4xc2.ent.gz pdb4xc2.ent.gz | 92.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4xc2.json.gz 4xc2.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/xc/4xc2 https://data.pdbj.org/pub/pdb/validation_reports/xc/4xc2 ftp://data.pdbj.org/pub/pdb/validation_reports/xc/4xc2 ftp://data.pdbj.org/pub/pdb/validation_reports/xc/4xc2 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  3d32S S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 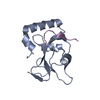
| ||||||||
| 2 | 
| ||||||||
| 3 | 
| ||||||||
| 4 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 13827.839 Da / Num. of mol.: 4 / Fragment: UNP residues 3-116 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: GABARAP, hCG_1987397 / Production host: Homo sapiens (human) / Gene: GABARAP, hCG_1987397 / Production host:   Escherichia coli (E. coli) / References: UniProt: Q6IAW1, UniProt: O95166*PLUS Escherichia coli (E. coli) / References: UniProt: Q6IAW1, UniProt: O95166*PLUS#2: Protein/peptide | Mass: 1307.388 Da / Num. of mol.: 4 / Fragment: UNP residues 663-673 / Source method: obtained synthetically / Source: (synth.)   Homo sapiens (human) / References: UniProt: Q86V97 Homo sapiens (human) / References: UniProt: Q86V97#3: Water | ChemComp-HOH / |  Water Water |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.19 Å3/Da / Density % sol: 43.75 % |
|---|---|
Crystal grow | Temperature: 293 K / Method: vapor diffusion, sitting drop / pH: 8 Details: 0.2 M Ammonium acetate, 0.01 M Magnesium acetate tetrahydrate, 30% Poly ethylene glycol 8000, 0.1 M Tris-HCl, pH 8.0 |
-Data collection
| Diffraction | Mean temperature: 77 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SLS SLS  / Beamline: X06SA / Wavelength: 1 Å / Beamline: X06SA / Wavelength: 1 Å |
| Detector | Type: PSI PILATUS 6M / Detector: PIXEL / Date: Jul 14, 2014 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 1 Å / Relative weight: 1 : 1 Å / Relative weight: 1 |
| Reflection | Resolution: 1.9→37.75 Å / Num. obs: 37496 / % possible obs: 92.7 % / Redundancy: 2.5 % / Rmerge(I) obs: 0.043 / Net I/σ(I): 12.1 |
| Reflection shell | Resolution: 1.9→2 Å / Redundancy: 2.4 % / Rmerge(I) obs: 0.365 / Mean I/σ(I) obs: 2.5 / Num. measured obs: 13090 / Num. unique all: 5380 / % possible all: 91 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 3D32 Resolution: 1.9→32.73 Å / Cor.coef. Fo:Fc: 0.966 / Cor.coef. Fo:Fc free: 0.94 / SU B: 5.492 / SU ML: 0.153 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.197 / ESU R Free: 0.181 / Stereochemistry target values: MAXIMUM LIKELIHOOD Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS U VALUES : REFINED INDIVIDUALLY
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 97.94 Å2 / Biso mean: 33.186 Å2 / Biso min: 12.66 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 1.9→32.73 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.9→1.949 Å / Total num. of bins used: 20
|
 Movie
Movie Controller
Controller













 PDBj
PDBj







