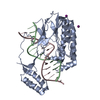
Entry Database : PDB / ID : 4s0hTitle TBX5 DB, NKX2.5 HD, ANF DNA Complex 5'-D(*CP*CP*AP*CP*TP*TP*CP*AP*AP*AP*GP*GP*TP*GP*TP*GP*AP*GP*A)-3'5'-D(*TP*CP*TP*CP*AP*CP*AP*CP*CP*TP*TP*TP*GP*AP*AP*GP*TP*GP*G)-3'Homeobox protein Nkx-2.5 T-box transcription factor TBX5 Keywords / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Homo sapiens (human)synthetic construct (others) Method / / / Resolution : 2.817 Å Authors Pradhan, L. Journal : Biochemistry / Year : 2016Title : Intermolecular Interactions of Cardiac Transcription Factors NKX2.5 and TBX5.Authors : Pradhan, L. / Gopal, S. / Li, S. / Ashur, S. / Suryanarayanan, S. / Kasahara, H. / Nam, H.J. History Deposition Dec 31, 2014 Deposition site / Processing site Revision 1.0 Dec 16, 2015 Provider / Type Revision 1.1 Mar 16, 2016 Group Revision 1.2 Apr 13, 2016 Group Revision 1.3 Feb 28, 2024 Group / Database referencesCategory chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / struct_ref_seq_dif Item / _database_2.pdbx_database_accession / _struct_ref_seq_dif.details
Show all Show less
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Transcription Factor / TRANSCRIPTION-DNA complex
Transcription Factor / TRANSCRIPTION-DNA complex Function and homology information
Function and homology information cardiac conduction system development / YAP1- and WWTR1 (TAZ)-stimulated gene expression / regulation of atrial cardiac muscle cell membrane depolarization / pattern specification process / negative regulation of epithelial to mesenchymal transition / pharyngeal system development / positive regulation of sodium ion transport / negative regulation of epithelial cell apoptotic process / outflow tract septum morphogenesis / cardiac muscle tissue morphogenesis / ventricular trabecula myocardium morphogenesis / heart trabecula formation / adult heart development / embryonic heart tube development / embryonic forelimb morphogenesis / aortic valve morphogenesis / cardiac muscle cell development / embryonic limb morphogenesis / Cardiogenesis / cell fate specification / negative regulation of cardiac muscle cell apoptotic process / atrioventricular valve morphogenesis / cardiac muscle cell proliferation / epithelial cell apoptotic process / ventricular septum development / DNA-binding transcription activator activity / ventricular septum morphogenesis / heart looping / cardiac septum morphogenesis / positive regulation of gap junction assembly / thyroid gland development /
cardiac conduction system development / YAP1- and WWTR1 (TAZ)-stimulated gene expression / regulation of atrial cardiac muscle cell membrane depolarization / pattern specification process / negative regulation of epithelial to mesenchymal transition / pharyngeal system development / positive regulation of sodium ion transport / negative regulation of epithelial cell apoptotic process / outflow tract septum morphogenesis / cardiac muscle tissue morphogenesis / ventricular trabecula myocardium morphogenesis / heart trabecula formation / adult heart development / embryonic heart tube development / embryonic forelimb morphogenesis / aortic valve morphogenesis / cardiac muscle cell development / embryonic limb morphogenesis / Cardiogenesis / cell fate specification / negative regulation of cardiac muscle cell apoptotic process / atrioventricular valve morphogenesis / cardiac muscle cell proliferation / epithelial cell apoptotic process / ventricular septum development / DNA-binding transcription activator activity / ventricular septum morphogenesis / heart looping / cardiac septum morphogenesis / positive regulation of gap junction assembly / thyroid gland development /  hemopoiesis / regulation of cardiac conduction / positive regulation of transcription initiation by RNA polymerase II / regulation of cardiac muscle contraction /
hemopoiesis / regulation of cardiac conduction / positive regulation of transcription initiation by RNA polymerase II / regulation of cardiac muscle contraction /  vasculogenesis / heart morphogenesis / positive regulation of cardiac muscle cell proliferation / cardiac muscle contraction / spleen development / epithelial cell differentiation / positive regulation of neuron differentiation / negative regulation of cell migration / epithelial cell proliferation / positive regulation of epithelial cell proliferation /
vasculogenesis / heart morphogenesis / positive regulation of cardiac muscle cell proliferation / cardiac muscle contraction / spleen development / epithelial cell differentiation / positive regulation of neuron differentiation / negative regulation of cell migration / epithelial cell proliferation / positive regulation of epithelial cell proliferation /  protein-DNA complex / morphogenesis of an epithelium / lung development / negative regulation of canonical Wnt signaling pathway / RNA polymerase II transcription regulator complex / sequence-specific double-stranded DNA binding / cell-cell signaling /
protein-DNA complex / morphogenesis of an epithelium / lung development / negative regulation of canonical Wnt signaling pathway / RNA polymerase II transcription regulator complex / sequence-specific double-stranded DNA binding / cell-cell signaling /  heart development / DNA-binding transcription activator activity, RNA polymerase II-specific /
heart development / DNA-binding transcription activator activity, RNA polymerase II-specific /  transcription regulator complex / RNA polymerase II-specific DNA-binding transcription factor binding / sequence-specific DNA binding / transcription by RNA polymerase II /
transcription regulator complex / RNA polymerase II-specific DNA-binding transcription factor binding / sequence-specific DNA binding / transcription by RNA polymerase II /  cell differentiation / transcription cis-regulatory region binding / DNA-binding transcription factor activity, RNA polymerase II-specific / RNA polymerase II cis-regulatory region sequence-specific DNA binding / DNA-binding transcription factor activity / negative regulation of cell population proliferation
cell differentiation / transcription cis-regulatory region binding / DNA-binding transcription factor activity, RNA polymerase II-specific / RNA polymerase II cis-regulatory region sequence-specific DNA binding / DNA-binding transcription factor activity / negative regulation of cell population proliferation
 Homo sapiens (human)
Homo sapiens (human) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.817 Å
MOLECULAR REPLACEMENT / Resolution: 2.817 Å  Authors
Authors Citation
Citation Journal: Biochemistry / Year: 2016
Journal: Biochemistry / Year: 2016 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 4s0h.cif.gz
4s0h.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb4s0h.ent.gz
pdb4s0h.ent.gz PDB format
PDB format 4s0h.json.gz
4s0h.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/s0/4s0h
https://data.pdbj.org/pub/pdb/validation_reports/s0/4s0h ftp://data.pdbj.org/pub/pdb/validation_reports/s0/4s0h
ftp://data.pdbj.org/pub/pdb/validation_reports/s0/4s0h Links
Links Assembly
Assembly


 Components
Components
 Homo sapiens (human) / Gene: TBX5 / Production host: unidentified (others) / References: UniProt: Q99593
Homo sapiens (human) / Gene: TBX5 / Production host: unidentified (others) / References: UniProt: Q99593 / Cardiac-specific homeobox / Homeobox protein CSX / Homeobox protein NK-2 homolog E
/ Cardiac-specific homeobox / Homeobox protein CSX / Homeobox protein NK-2 homolog E
 Homo sapiens (human) / Gene: NKX2-5, CSX, NKX2.5, NKX2E / Production host: unidentified (others) / References: UniProt: P52952
Homo sapiens (human) / Gene: NKX2-5, CSX, NKX2.5, NKX2E / Production host: unidentified (others) / References: UniProt: P52952 Water
Water X-RAY DIFFRACTION
X-RAY DIFFRACTION Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  CHESS
CHESS  / Beamline: A1 / Wavelength: 0.9767 Å
/ Beamline: A1 / Wavelength: 0.9767 Å : 0.9767 Å / Relative weight: 1
: 0.9767 Å / Relative weight: 1  Processing
Processing :
:  MOLECULAR REPLACEMENT / Resolution: 2.817→38.164 Å / SU ML: 0.38 / σ(F): 0.12 / Phase error: 28.93 / Stereochemistry target values: MLHL
MOLECULAR REPLACEMENT / Resolution: 2.817→38.164 Å / SU ML: 0.38 / σ(F): 0.12 / Phase error: 28.93 / Stereochemistry target values: MLHL Movie
Movie Controller
Controller













 PDBj
PDBj










































