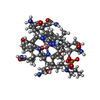
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4frg | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of the cobalamin riboswitch aptamer domain | ||||||
 Components Components | cobalamin riboswitch aptamer domain | ||||||
 Keywords Keywords |  RNA / RNA /  cobalamin / cobalamin /  riboswitch / B12 riboswitch / B12 | ||||||
| Function / homology |  Hydroxocobalamin / IRIDIUM (III) ION / Hydroxocobalamin / IRIDIUM (III) ION /  RNA / RNA (> 10) RNA / RNA (> 10) Function and homology information Function and homology information | ||||||
| Biological species | Marine metagenome (others) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MAD / Resolution: 2.95 Å MAD / Resolution: 2.95 Å | ||||||
 Authors Authors | Reyes, F.E. / Johnson, J.E. / Polaski, J.T. / Batey, R.T. | ||||||
 Citation Citation |  Journal: Nature / Year: 2012 Journal: Nature / Year: 2012Title: B12 cofactors directly stabilize an mRNA regulatory switch. Authors: Johnson, J.E. / Reyes, F.E. / Polaski, J.T. / Batey, R.T. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4frg.cif.gz 4frg.cif.gz | 196.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4frg.ent.gz pdb4frg.ent.gz | 160.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4frg.json.gz 4frg.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/fr/4frg https://data.pdbj.org/pub/pdb/validation_reports/fr/4frg ftp://data.pdbj.org/pub/pdb/validation_reports/fr/4frg ftp://data.pdbj.org/pub/pdb/validation_reports/fr/4frg | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: RNA chain | Mass: 27135.234 Da / Num. of mol.: 2 / Source method: obtained synthetically / Details: RNA was transcribed via in vitro T7 RNA polymerase / Source: (synth.) Marine metagenome (others) #2: Chemical |  Hydroxocobalamin Hydroxocobalamin#3: Chemical | ChemComp-IR3 /  Iridium Iridium#4: Chemical | ChemComp-MG / Sequence details | THE SEQUENCE CAN BE FOUND AT GENBANK ACCESSION #: AACY021350931.1/557-477 OBTAINED FROM SHOTGUN ...THE SEQUENCE CAN BE FOUND AT GENBANK ACCESSION #: AACY021350931.1/557-477 OBTAINED FROM SHOTGUN SEQUENCING | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal |
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
Crystal grow |
|
-Data collection
| Diffraction |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Detector | Type: ADSC QUANTUM 315 / Detector: CCD / Date: Dec 12, 2010 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation | Protocol: MAD / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation wavelength |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection | Redundancy: 13.5 % / Av σ(I) over netI: 6.8 / Number: 192234 / Rsym value: 0.082 / D res high: 3.16 Å / D res low: 137.897 Å / Num. obs: 14220 / % possible obs: 97.2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Diffraction reflection shell |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection | Resolution: 2.95→137.899 Å / Num. all: 17854 / Num. obs: 17854 / % possible obs: 99.9 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Redundancy: 13.7 % / Rsym value: 0.098 / Net I/σ(I): 20.6 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection shell | Rmerge(I) obs: 0.014 / Diffraction-ID: 1
|
-Phasing
Phasing | Method:  MAD MAD | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Phasing MAD | D res high: 3.16 Å / D res low: 43.06 Å / FOM acentric: 0.358 / FOM centric: 0.227 / Reflection acentric: 11936 / Reflection centric: 2423 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Phasing MAD set | Highest resolution: 3.16 Å / Lowest resolution: 43.06 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Phasing MAD set shell |
|
 Movie
Movie Controller
Controller














 PDBj
PDBj