+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3wav | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Crystal Structure of Autotaxin in Complex with Compound 10 | |||||||||
 Components Components | Ectonucleotide pyrophosphatase/phosphodiesterase family member 2 | |||||||||
 Keywords Keywords | HYDROLASE/HYDROLASE INHIBITOR / HYDROLASE-HYDROLASE INHIBITOR complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationdinucleotide phosphatase activity /  alkylglycerophosphoethanolamine phosphodiesterase / sphingolipid catabolic process / phospholipid catabolic process / phosphatidylcholine catabolic process / alkylglycerophosphoethanolamine phosphodiesterase / sphingolipid catabolic process / phospholipid catabolic process / phosphatidylcholine catabolic process /  lysophospholipase activity / positive regulation of lamellipodium morphogenesis / lysophospholipase activity / positive regulation of lamellipodium morphogenesis /  phosphodiesterase I activity / scavenger receptor activity / phosphodiesterase I activity / scavenger receptor activity /  alkylglycerophosphoethanolamine phosphodiesterase activity ...dinucleotide phosphatase activity / alkylglycerophosphoethanolamine phosphodiesterase activity ...dinucleotide phosphatase activity /  alkylglycerophosphoethanolamine phosphodiesterase / sphingolipid catabolic process / phospholipid catabolic process / phosphatidylcholine catabolic process / alkylglycerophosphoethanolamine phosphodiesterase / sphingolipid catabolic process / phospholipid catabolic process / phosphatidylcholine catabolic process /  lysophospholipase activity / positive regulation of lamellipodium morphogenesis / lysophospholipase activity / positive regulation of lamellipodium morphogenesis /  phosphodiesterase I activity / scavenger receptor activity / phosphodiesterase I activity / scavenger receptor activity /  alkylglycerophosphoethanolamine phosphodiesterase activity / alkylglycerophosphoethanolamine phosphodiesterase activity /  polysaccharide binding / positive regulation of oligodendrocyte differentiation / negative regulation of cell-matrix adhesion / positive regulation of epithelial cell migration / positive regulation of focal adhesion assembly / polysaccharide binding / positive regulation of oligodendrocyte differentiation / negative regulation of cell-matrix adhesion / positive regulation of epithelial cell migration / positive regulation of focal adhesion assembly /  estrous cycle / phospholipid metabolic process / positive regulation of substrate adhesion-dependent cell spreading / estrous cycle / phospholipid metabolic process / positive regulation of substrate adhesion-dependent cell spreading /  regulation of cell migration / cell chemotaxis / positive regulation of peptidyl-tyrosine phosphorylation / regulation of cell migration / cell chemotaxis / positive regulation of peptidyl-tyrosine phosphorylation /  nucleic acid binding / nucleic acid binding /  immune response / immune response /  calcium ion binding / positive regulation of cell population proliferation / calcium ion binding / positive regulation of cell population proliferation /  extracellular space / zinc ion binding / extracellular space / zinc ion binding /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |   Mus musculus (house mouse) Mus musculus (house mouse) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.797 Å MOLECULAR REPLACEMENT / Resolution: 1.797 Å | |||||||||
 Authors Authors | Nishimasu, H. / Ishitani, R. / Nureki, O. | |||||||||
 Citation Citation |  Journal: Acs Chem.Biol. / Year: 2013 Journal: Acs Chem.Biol. / Year: 2013Title: Screening and X-ray Crystal Structure-based Optimization of Autotaxin (ENPP2) Inhibitors, Using a Newly Developed Fluorescence Probe Authors: Kawaguchi, M. / Okabe, T. / Okudaira, S. / Nishimasu, H. / Ishitani, R. / Kojima, H. / Nureki, O. / Aoki, J. / Nagano, T. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3wav.cif.gz 3wav.cif.gz | 196.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3wav.ent.gz pdb3wav.ent.gz | 150.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3wav.json.gz 3wav.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/wa/3wav https://data.pdbj.org/pub/pdb/validation_reports/wa/3wav ftp://data.pdbj.org/pub/pdb/validation_reports/wa/3wav ftp://data.pdbj.org/pub/pdb/validation_reports/wa/3wav | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  3wawC  3waxC  3wayC  3nkmS C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 1 molecules A
| #1: Protein | Mass: 95757.430 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Mus musculus (house mouse) / Gene: Enpp2 / Plasmid: pCD-CW / Cell line (production host): HEK293S GnT1- / Production host: Mus musculus (house mouse) / Gene: Enpp2 / Plasmid: pCD-CW / Cell line (production host): HEK293S GnT1- / Production host:   Homo Sapiens (human) Homo Sapiens (human)References: UniProt: Q9R1E6,  alkylglycerophosphoethanolamine phosphodiesterase alkylglycerophosphoethanolamine phosphodiesterase |
|---|
-Sugars , 2 types, 3 molecules
| #2: Polysaccharide |  / Mass: 424.401 Da / Num. of mol.: 2 / Mass: 424.401 Da / Num. of mol.: 2Source method: isolated from a genetically manipulated source #3: Polysaccharide | alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-beta-D- ...alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose |  / Mass: 1072.964 Da / Num. of mol.: 1 / Mass: 1072.964 Da / Num. of mol.: 1Source method: isolated from a genetically manipulated source |
|---|
-Non-polymers , 9 types, 450 molecules 






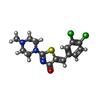









| #4: Chemical | | #5: Chemical | ChemComp-CA / | #6: Chemical | ChemComp-NA / | #7: Chemical | ChemComp-K / | #8: Chemical |  Thiocyanate Thiocyanate#9: Chemical | ChemComp-EDO /  Ethylene glycol Ethylene glycol#10: Chemical | ChemComp-SO4 / |  Sulfate Sulfate#11: Chemical | ChemComp-DWV / ( | #12: Water | ChemComp-HOH / |  Water Water |
|---|
-Details
| Sequence details | PROTEIN USED IN THIS STRUCTURE IS AN ISOFORM OF AUTOTAXIN FROM MOUSE, WHICH IS LACK OF RESIDUES ...PROTEIN USED IN THIS STRUCTURE IS AN ISOFORM OF AUTOTAXIN FROM MOUSE, WHICH IS LACK OF RESIDUES KVEP (UNP RESDIUES 571-574 OF DATABASE ENPP2_MOUSE). |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.27 Å3/Da / Density % sol: 45.83 % |
|---|---|
Crystal grow | Temperature: 293 K / Method: vapor diffusion, sitting drop Details: 23% PEG3350, 0.15M NaCl, 0.5M KSCN, 0.2mM ZnSO4, 1% polyvinylpyrrolidone, vapor diffusion, sitting drop, temperature 293K |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SPring-8 SPring-8  / Beamline: BL41XU / Wavelength: 1 Å / Beamline: BL41XU / Wavelength: 1 Å |
| Detector | Detector: CCD / Date: Apr 21, 2010 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 1 Å / Relative weight: 1 : 1 Å / Relative weight: 1 |
| Reflection | Resolution: 1.797→50 Å / Num. obs: 77258 / Biso Wilson estimate: 25.6 Å2 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 3NKM Resolution: 1.797→33.245 Å / Occupancy max: 1 / Occupancy min: 0.5 / FOM work R set: 0.8569 / SU ML: 0.47 / σ(F): 1.43 / Phase error: 22.37 / Stereochemistry target values: ML
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 1.11 Å / VDW probe radii: 1.3 Å / Solvent model: FLAT BULK SOLVENT MODEL / Bsol: 44.055 Å2 / ksol: 0.349 e/Å3 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 87.52 Å2 / Biso mean: 35.4461 Å2 / Biso min: 11.38 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.797→33.245 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Total num. of bins used: 28
|
 Movie
Movie Controller
Controller













 PDBj
PDBj









