[English] 日本語
 Yorodumi
Yorodumi- PDB-3v33: Crystal structure of MCPIP1 conserved domain with zinc-finger motif -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3v33 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of MCPIP1 conserved domain with zinc-finger motif | ||||||
 Components Components | Ribonuclease ZC3H12A | ||||||
 Keywords Keywords |  HYDROLASE / Rossmann-like sandwich fold / HYDROLASE / Rossmann-like sandwich fold /  RNase / cytoplastic RNase / cytoplastic | ||||||
| Function / homology |  Function and homology information Function and homology informationimmune response-activating signaling pathway / positive regulation of miRNA catabolic process / positive regulation of protein deubiquitination / cellular response to ionomycin / miRNA catabolic process / Regulation of CDH19 Expression and Function / negative regulation of macrophage activation / RNA exonuclease activity / positive regulation of mRNA catabolic process / rough endoplasmic reticulum membrane ...immune response-activating signaling pathway / positive regulation of miRNA catabolic process / positive regulation of protein deubiquitination / cellular response to ionomycin / miRNA catabolic process / Regulation of CDH19 Expression and Function / negative regulation of macrophage activation / RNA exonuclease activity / positive regulation of mRNA catabolic process / rough endoplasmic reticulum membrane / negative regulation by host of viral genome replication / cellular response to sodium arsenite / negative regulation of T-helper 17 cell differentiation / negative regulation of muscle cell apoptotic process / 3'-UTR-mediated mRNA destabilization / positive regulation of lipid storage / negative regulation of cardiac muscle contraction / negative regulation of nitric oxide biosynthetic process / mRNA 3'-UTR AU-rich region binding / cellular response to chemokine / negative regulation of non-canonical NF-kappaB signal transduction / miRNA binding / nuclear-transcribed mRNA catabolic process, nonsense-mediated decay / negative regulation of interleukin-1 beta production / positive regulation of p38MAPK cascade / positive regulation of execution phase of apoptosis / negative regulation of NF-kappaB transcription factor activity / protein deubiquitination / negative regulation of interleukin-6 production / negative regulation of type II interferon production / negative regulation of tumor necrosis factor production / negative regulation of cytokine production involved in inflammatory response / positive regulation of fat cell differentiation / cellular response to interleukin-1 / RNA nuclease activity / cellular response to glucose starvation / positive regulation of autophagy / RNA stem-loop binding / negative regulation of canonical NF-kappaB signal transduction / RNA endonuclease activity / positive regulation of defense response to virus by host / positive regulation of endothelial cell migration / negative regulation of protein phosphorylation / mRNA 3'-UTR binding /  P-body / cellular response to virus / positive regulation of protein import into nucleus / cytoplasmic ribonucleoprotein granule / positive regulation of angiogenesis / positive regulation of reactive oxygen species metabolic process / P-body / cellular response to virus / positive regulation of protein import into nucleus / cytoplasmic ribonucleoprotein granule / positive regulation of angiogenesis / positive regulation of reactive oxygen species metabolic process /  ribosome binding / protein complex oligomerization / cellular response to tumor necrosis factor / ribosome binding / protein complex oligomerization / cellular response to tumor necrosis factor /  nervous system development / cellular response to oxidative stress / T cell receptor signaling pathway / nervous system development / cellular response to oxidative stress / T cell receptor signaling pathway /  regulation of gene expression / regulation of gene expression /  angiogenesis / defense response to virus / cellular response to lipopolysaccharide / angiogenesis / defense response to virus / cellular response to lipopolysaccharide /  Hydrolases; Acting on ester bonds / Hydrolases; Acting on ester bonds /  cell differentiation / cell differentiation /  cytoskeleton / cytoskeleton /  inflammatory response / negative regulation of gene expression / inflammatory response / negative regulation of gene expression /  mRNA binding / apoptotic process / DNA damage response / mRNA binding / apoptotic process / DNA damage response /  chromatin binding / positive regulation of gene expression / positive regulation of transcription by RNA polymerase II / protein-containing complex / chromatin binding / positive regulation of gene expression / positive regulation of transcription by RNA polymerase II / protein-containing complex /  DNA binding / DNA binding /  RNA binding / RNA binding /  nucleoplasm / nucleoplasm /  metal ion binding / metal ion binding /  nucleus / nucleus /  cytoplasm cytoplasmSimilarity search - Function | ||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.005 Å MOLECULAR REPLACEMENT / Resolution: 2.005 Å | ||||||
 Authors Authors | Xu, J. / Peng, W. / Sun, Y. / Wang, X. / Xu, Y. / Li, X. / Gao, G. / Rao, Z. | ||||||
 Citation Citation |  Journal: Nucleic Acids Res. / Year: 2012 Journal: Nucleic Acids Res. / Year: 2012Title: Structural study of MCPIP1 N-terminal conserved domain reveals a PIN-like RNase Authors: Xu, J. / Peng, W. / Sun, Y. / Wang, X. / Xu, Y. / Li, X. / Gao, G. / Rao, Z. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3v33.cif.gz 3v33.cif.gz | 147.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3v33.ent.gz pdb3v33.ent.gz | 117.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3v33.json.gz 3v33.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/v3/3v33 https://data.pdbj.org/pub/pdb/validation_reports/v3/3v33 ftp://data.pdbj.org/pub/pdb/validation_reports/v3/3v33 ftp://data.pdbj.org/pub/pdb/validation_reports/v3/3v33 | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 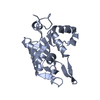
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
| ||||||||
| Components on special symmetry positions |
|
- Components
Components
| #1: Protein | Mass: 25772.594 Da / Num. of mol.: 2 Fragment: N-terminal conserved domain with the zinc-finger motif, residues 112-334 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: MCPIP1 / Plasmid: pGEX 6p-1 / Production host: Homo sapiens (human) / Gene: MCPIP1 / Plasmid: pGEX 6p-1 / Production host:   Escherichia coli (E. coli) / Strain (production host): BL21(DE3) Escherichia coli (E. coli) / Strain (production host): BL21(DE3)References: UniProt: Q5D1E8,  Hydrolases; Acting on ester bonds Hydrolases; Acting on ester bonds#2: Water | ChemComp-HOH / |  Water Water |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.82 Å3/Da / Density % sol: 56.32 % |
|---|---|
Crystal grow | Temperature: 289 K / Method: vapor diffusion, hanging drop / pH: 5 Details: 4% Tacsimate, 6% PEG 3350, VAPOR DIFFUSION, HANGING DROP, temperature 289K |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SSRF SSRF  / Beamline: BL17U / Wavelength: 1 Å / Beamline: BL17U / Wavelength: 1 Å |
| Detector | Type: RAYONIX MX-225 / Detector: CCD / Date: Jul 13, 2010 |
| Radiation | Monochromator: SAGITALLY FOCUSED Si(111) / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 1 Å / Relative weight: 1 : 1 Å / Relative weight: 1 |
| Reflection | Resolution: 2→36.26 Å / Num. all: 39295 / Num. obs: 30650 / % possible obs: 78 % / Observed criterion σ(F): 1 / Observed criterion σ(I): 1 / Redundancy: 5.8 % / Biso Wilson estimate: 32.81 Å2 / Rmerge(I) obs: 0.086 / Net I/σ(I): 26.2 |
| Reflection shell | Resolution: 2→2.07 Å / Redundancy: 2.1 % / Rmerge(I) obs: 0.567 / Mean I/σ(I) obs: 1.35 / % possible all: 33.4 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT / Resolution: 2.005→36.259 Å / Occupancy max: 1 / Occupancy min: 0 / FOM work R set: 0.7982 / SU ML: 0.24 / σ(F): 1.34 / Phase error: 26.87 / Stereochemistry target values: ML MOLECULAR REPLACEMENT / Resolution: 2.005→36.259 Å / Occupancy max: 1 / Occupancy min: 0 / FOM work R set: 0.7982 / SU ML: 0.24 / σ(F): 1.34 / Phase error: 26.87 / Stereochemistry target values: ML
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL / Bsol: 33.605 Å2 / ksol: 0.332 e/Å3 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 141.18 Å2 / Biso mean: 47.5958 Å2 / Biso min: 12.34 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.005→36.259 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Total num. of bins used: 10
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Origin x: -58.4348 Å / Origin y: 0.746 Å / Origin z: 10.4629 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller














 PDBj
PDBj
