[English] 日本語
 Yorodumi
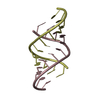
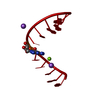
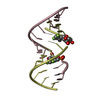
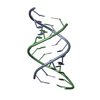
Yorodumi- PDB-3ssf: Crystal structure of RNA:DNA dodecamer corresponding to HIV-1 pol... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3ssf | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of RNA:DNA dodecamer corresponding to HIV-1 polypurine tract, at 1.6 A resolution. | ||||||
 Components Components |
| ||||||
 Keywords Keywords | RNA/DNA / Polypurine tract (PPT) of HIV-1 / RNA-DNA hybrid /  reverse transcription / RNA-DNA complex reverse transcription / RNA-DNA complex | ||||||
| Function / homology |  DNA / DNA (> 10) / DNA / DNA (> 10) /  RNA / RNA (> 10) RNA / RNA (> 10) Function and homology information Function and homology information | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.6 Å MOLECULAR REPLACEMENT / Resolution: 1.6 Å | ||||||
 Authors Authors | Drozdzal, P. / Michalska, K. / Kierzek, R. / Lomozik, L. / Jaskolski, M. | ||||||
 Citation Citation |  Journal: Acta Crystallogr.,Sect.D / Year: 2012 Journal: Acta Crystallogr.,Sect.D / Year: 2012Title: Structure of an RNA/DNA dodecamer corresponding to the HIV-1 polypurine tract at 1.6 Angstrom resolution Authors: Drozdzal, P. / Michalska, K. / Kierzek, R. / Lomozik, L. / Jaskolski, M. #1:  Journal: J.Mol.Biol. / Year: 2003 Journal: J.Mol.Biol. / Year: 2003Title: An unusual sugar conformation in the structure of an RNA/DNA decamer of the polypurine tract may affect recognition by RNase H. Authors: Kopka, M.L. / Lavelle, L. / Han, G.W. / Ng, H.-L. / Dickerson, R.E. #2:  Journal: Proc.Natl.Acad.Sci.USA / Year: 2003 Journal: Proc.Natl.Acad.Sci.USA / Year: 2003Title: Crystal structure of an RNA.DNA hybrid reveals intermolecular intercalation: dimer formation by base-pair swapping. Authors: Han, G.W. / Kopka, M.L. / Langs, D. / Sawaya, M.R. / Dickerson, R.E. #3:  Journal: Embo J. / Year: 2001 Journal: Embo J. / Year: 2001Title: Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA Authors: G Sarafianos, S. / Das, K. / Tantillo, C. / Clark Jr., A.D. / Ding, J. / Whitcomb, J.M. / Boyer, P.L. / Hughes, S.H. / Arnold, E. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3ssf.cif.gz 3ssf.cif.gz | 43.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3ssf.ent.gz pdb3ssf.ent.gz | 31.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3ssf.json.gz 3ssf.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ss/3ssf https://data.pdbj.org/pub/pdb/validation_reports/ss/3ssf ftp://data.pdbj.org/pub/pdb/validation_reports/ss/3ssf ftp://data.pdbj.org/pub/pdb/validation_reports/ss/3ssf | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1pjoS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Components on special symmetry positions |
|
- Components
Components
| #1: RNA chain | Mass: 3930.472 Da / Num. of mol.: 1 / Source method: obtained synthetically | ||
|---|---|---|---|
| #2: DNA chain | Mass: 3569.337 Da / Num. of mol.: 1 / Source method: obtained synthetically | ||
| #3: Chemical | | #4: Water | ChemComp-HOH / |  Water Water |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 1.94 Å3/Da / Density % sol: 36.7 % |
|---|---|
Crystal grow | Temperature: 292 K / pH: 5.5 Details: 40% MPD, 0.04M hexamminecobalt(III) chloride, 0.02M magnesium chloride, 0.04M lithium chloride, 0.04M sodium cacodylate, pH 5.5, VAPOR DIFFUSION, HANGING DROP, temperature 292K |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  EMBL/DESY, HAMBURG EMBL/DESY, HAMBURG  / Beamline: X13 / Wavelength: 0.801 / Beamline: X13 / Wavelength: 0.801 |
| Detector | Type: MAR CCD 165 mm / Detector: CCD / Date: Dec 4, 2009 |
| Radiation | Monochromator: SI 111 CHANNEL / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 0.801 Å / Relative weight: 1 : 0.801 Å / Relative weight: 1 |
| Reflection | Resolution: 1.6→50 Å / Num. obs: 8012 / % possible obs: 99.5 % / Observed criterion σ(I): -3 / Redundancy: 6.7 % / Rmerge(I) obs: 0.04 / Net I/σ(I): 41.7 |
| Reflection shell | Resolution: 1.6→1.66 Å / Redundancy: 5.5 % / Rmerge(I) obs: 0.55 / Mean I/σ(I) obs: 2.851 / % possible all: 99.6 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 1PJO Resolution: 1.6→15.32 Å / Cor.coef. Fo:Fc: 0.972 / Cor.coef. Fo:Fc free: 0.958 / SU B: 4.319 / SU ML: 0.075 / Cross valid method: THROUGHOUT / ESU R: 0.108 / ESU R Free: 0.111 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGEN ATOMS WERE ADDED IN THE RIDING POSITIONS
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.4 Å / Solvent model: BABINET MODEL WITH MASK | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 23.02 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.6→15.32 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.6→1.64 Å / Total num. of bins used: 20
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller





 PDBj
PDBj


































































