+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3sh5 | ||||||
|---|---|---|---|---|---|---|---|
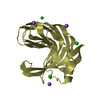
| Title | Calcium-bound Laminin G like domain 3 from human perlecan | ||||||
 Components Components | LG3 peptide | ||||||
 Keywords Keywords | METAL BINDING PROTEIN / Actin disassambly activity / integrin alpha 2 beta 1 | ||||||
| Function / homology |  Function and homology information Function and homology informationextracellular matrix structural constituent conferring compression resistance /  collagen V binding / Defective B3GALT6 causes EDSP2 and SEMDJL1 / Defective B4GALT7 causes EDS, progeroid type / Defective B3GAT3 causes JDSSDHD / Defective EXT2 causes exostoses 2 / Defective EXT1 causes exostoses 1, TRPS2 and CHDS / A tetrasaccharide linker sequence is required for GAG synthesis / HS-GAG biosynthesis / HS-GAG degradation ...extracellular matrix structural constituent conferring compression resistance / collagen V binding / Defective B3GALT6 causes EDSP2 and SEMDJL1 / Defective B4GALT7 causes EDS, progeroid type / Defective B3GAT3 causes JDSSDHD / Defective EXT2 causes exostoses 2 / Defective EXT1 causes exostoses 1, TRPS2 and CHDS / A tetrasaccharide linker sequence is required for GAG synthesis / HS-GAG biosynthesis / HS-GAG degradation ...extracellular matrix structural constituent conferring compression resistance /  collagen V binding / Defective B3GALT6 causes EDSP2 and SEMDJL1 / Defective B4GALT7 causes EDS, progeroid type / Defective B3GAT3 causes JDSSDHD / Defective EXT2 causes exostoses 2 / Defective EXT1 causes exostoses 1, TRPS2 and CHDS / A tetrasaccharide linker sequence is required for GAG synthesis / HS-GAG biosynthesis / HS-GAG degradation / collagen V binding / Defective B3GALT6 causes EDSP2 and SEMDJL1 / Defective B4GALT7 causes EDS, progeroid type / Defective B3GAT3 causes JDSSDHD / Defective EXT2 causes exostoses 2 / Defective EXT1 causes exostoses 1, TRPS2 and CHDS / A tetrasaccharide linker sequence is required for GAG synthesis / HS-GAG biosynthesis / HS-GAG degradation /  circulatory system development / Laminin interactions / circulatory system development / Laminin interactions /  plasma membrane protein complex / negative regulation of cell adhesion / smoothened signaling pathway / low-density lipoprotein particle receptor binding / Non-integrin membrane-ECM interactions / plasma membrane protein complex / negative regulation of cell adhesion / smoothened signaling pathway / low-density lipoprotein particle receptor binding / Non-integrin membrane-ECM interactions /  basement membrane / ECM proteoglycans / Integrin cell surface interactions / animal organ regeneration / Retinoid metabolism and transport / positive regulation of endothelial cell proliferation / basement membrane / ECM proteoglycans / Integrin cell surface interactions / animal organ regeneration / Retinoid metabolism and transport / positive regulation of endothelial cell proliferation /  embryo implantation / Degradation of the extracellular matrix / lysosomal lumen / embryo implantation / Degradation of the extracellular matrix / lysosomal lumen /  receptor-mediated endocytosis / negative regulation of angiogenesis / receptor-mediated endocytosis / negative regulation of angiogenesis /  brain development / lipid metabolic process / Golgi lumen / brain development / lipid metabolic process / Golgi lumen /  amyloid-beta binding / amyloid-beta binding /  angiogenesis / collagen-containing extracellular matrix / Attachment and Entry / angiogenesis / collagen-containing extracellular matrix / Attachment and Entry /  cell differentiation / response to hypoxia / response to xenobiotic stimulus / cell differentiation / response to hypoxia / response to xenobiotic stimulus /  inflammatory response / Amyloid fiber formation / negative regulation of cell population proliferation / inflammatory response / Amyloid fiber formation / negative regulation of cell population proliferation /  focal adhesion / focal adhesion /  calcium ion binding / calcium ion binding /  extracellular space / extracellular exosome / extracellular region / extracellular space / extracellular exosome / extracellular region /  plasma membrane plasma membraneSimilarity search - Function | ||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.8 Å MOLECULAR REPLACEMENT / Resolution: 2.8 Å | ||||||
 Authors Authors | Van Le, B. / Kim, K.K. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 2011 Journal: J.Mol.Biol. / Year: 2011Title: Crystal Structure of the LG3 Domain of Endorepellin, an Angiogenesis Inhibitor. Authors: Van Le, B. / Kim, H. / Choi, J. / Kim, J.H. / Hahn, M.J. / Lee, C. / Kim, K.K. / Hwang, H.Y. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3sh5.cif.gz 3sh5.cif.gz | 43.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3sh5.ent.gz pdb3sh5.ent.gz | 33 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3sh5.json.gz 3sh5.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/sh/3sh5 https://data.pdbj.org/pub/pdb/validation_reports/sh/3sh5 ftp://data.pdbj.org/pub/pdb/validation_reports/sh/3sh5 ftp://data.pdbj.org/pub/pdb/validation_reports/sh/3sh5 | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 20571.803 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Homo sapiens (human) / References: UniProt: P98160 Homo sapiens (human) / References: UniProt: P98160 |
|---|---|
| #2: Chemical | ChemComp-CA / |
| #3: Water | ChemComp-HOH /  Water Water |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 1.89 Å3/Da / Density % sol: 35.03 % |
|---|---|
Crystal grow | Temperature: 293 K / Method: vapor diffusion, hanging drop / pH: 4.5 Details: 20% PEG 8000, 200mM calcium acetate, 100mM MES pH 6.0, VAPOR DIFFUSION, HANGING DROP, temperature 293K |
-Data collection
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Photon Factory Photon Factory  / Beamline: AR-NW12A / Beamline: AR-NW12A |
|---|---|
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Relative weight: 1 |
| Reflection | Resolution: 2.8→29 Å / Num. all: 4115 / Num. obs: 4115 / % possible obs: 100 % / Observed criterion σ(F): 1 / Observed criterion σ(I): 3 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT / Resolution: 2.8→29 Å / Cor.coef. Fo:Fc: 0.911 / Cor.coef. Fo:Fc free: 0.85 / Rfactor Rfree error: 0.021 / Occupancy max: 1 / Occupancy min: 1 / SU B: 14.952 / SU ML: 0.313 / Data cutoff high absF: 1065743 / Isotropic thermal model: RESTRAINED / Cross valid method: THROUGHOUT / ESU R Free: 0.512 / Stereochemistry target values: MAXIMUM LIKELIHOOD MOLECULAR REPLACEMENT / Resolution: 2.8→29 Å / Cor.coef. Fo:Fc: 0.911 / Cor.coef. Fo:Fc free: 0.85 / Rfactor Rfree error: 0.021 / Occupancy max: 1 / Occupancy min: 1 / SU B: 14.952 / SU ML: 0.313 / Data cutoff high absF: 1065743 / Isotropic thermal model: RESTRAINED / Cross valid method: THROUGHOUT / ESU R Free: 0.512 / Stereochemistry target values: MAXIMUM LIKELIHOOD
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.4 Å / Solvent model: FLAT MODEL | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 56.41 Å2 / Biso mean: 22.383 Å2 / Biso min: 9.67 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.8→29 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.803→2.875 Å / Total num. of bins used: 20
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Xplor file |
|
 Movie
Movie Controller
Controller













 PDBj
PDBj




















