+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3s67 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of V57P mutant of human cystatin C | ||||||
 Components Components | Cystatin-C | ||||||
 Keywords Keywords | HYDROLASE INHIBITOR | ||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of collagen catabolic process / negative regulation of elastin catabolic process / negative regulation of blood vessel remodeling / negative regulation of peptidase activity /  peptidase inhibitor activity / negative regulation of extracellular matrix disassembly / peptidase inhibitor activity / negative regulation of extracellular matrix disassembly /  regulation of tissue remodeling / cysteine-type endopeptidase inhibitor activity / regulation of tissue remodeling / cysteine-type endopeptidase inhibitor activity /  endopeptidase inhibitor activity / supramolecular fiber organization ...negative regulation of collagen catabolic process / negative regulation of elastin catabolic process / negative regulation of blood vessel remodeling / negative regulation of peptidase activity / endopeptidase inhibitor activity / supramolecular fiber organization ...negative regulation of collagen catabolic process / negative regulation of elastin catabolic process / negative regulation of blood vessel remodeling / negative regulation of peptidase activity /  peptidase inhibitor activity / negative regulation of extracellular matrix disassembly / peptidase inhibitor activity / negative regulation of extracellular matrix disassembly /  regulation of tissue remodeling / cysteine-type endopeptidase inhibitor activity / regulation of tissue remodeling / cysteine-type endopeptidase inhibitor activity /  endopeptidase inhibitor activity / supramolecular fiber organization / negative regulation of proteolysis / endopeptidase inhibitor activity / supramolecular fiber organization / negative regulation of proteolysis /  Post-translational protein phosphorylation / defense response / Regulation of Insulin-like Growth Factor (IGF) transport and uptake by Insulin-like Growth Factor Binding Proteins (IGFBPs) / tertiary granule lumen / Post-translational protein phosphorylation / defense response / Regulation of Insulin-like Growth Factor (IGF) transport and uptake by Insulin-like Growth Factor Binding Proteins (IGFBPs) / tertiary granule lumen /  amyloid-beta binding / amyloid-beta binding /  protease binding / vesicle / ficolin-1-rich granule lumen / Amyloid fiber formation / protease binding / vesicle / ficolin-1-rich granule lumen / Amyloid fiber formation /  endoplasmic reticulum lumen / Neutrophil degranulation / endoplasmic reticulum lumen / Neutrophil degranulation /  Golgi apparatus / Golgi apparatus /  endoplasmic reticulum / endoplasmic reticulum /  extracellular space / extracellular exosome / extracellular region / identical protein binding / extracellular space / extracellular exosome / extracellular region / identical protein binding /  plasma membrane / plasma membrane /  cytoplasm cytoplasmSimilarity search - Function | ||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.26 Å MOLECULAR REPLACEMENT / Resolution: 2.26 Å | ||||||
 Authors Authors | Orlikowska, M. / Szymanska, A. / Borek, D. / Otwinowski, Z. / Skowron, P. / Jankowska, E. | ||||||
 Citation Citation |  Journal: Acta Crystallogr.,Sect.D / Year: 2013 Journal: Acta Crystallogr.,Sect.D / Year: 2013Title: Structural characterization of V57D and V57P mutants of human cystatin C, an amyloidogenic protein. Authors: Orlikowska, M. / Szymanska, A. / Borek, D. / Otwinowski, Z. / Skowron, P. / Jankowska, E. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3s67.cif.gz 3s67.cif.gz | 62.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3s67.ent.gz pdb3s67.ent.gz | 47.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3s67.json.gz 3s67.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/s6/3s67 https://data.pdbj.org/pub/pdb/validation_reports/s6/3s67 ftp://data.pdbj.org/pub/pdb/validation_reports/s6/3s67 ftp://data.pdbj.org/pub/pdb/validation_reports/s6/3s67 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  3svaC  1g96S S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 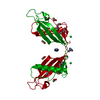
| |||||||||
| Unit cell |
| |||||||||
| Components on special symmetry positions |
|
- Components
Components
-Protein , 1 types, 1 molecules A
| #1: Protein | Mass: 13363.124 Da / Num. of mol.: 1 / Mutation: V57P Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: CST3 / Plasmid: pHD313 / Production host: Homo sapiens (human) / Gene: CST3 / Plasmid: pHD313 / Production host:   Escherichia coli (E. coli) / Strain (production host): C41 / References: UniProt: P01034 Escherichia coli (E. coli) / Strain (production host): C41 / References: UniProt: P01034 |
|---|
-Non-polymers , 5 types, 76 molecules 








| #2: Chemical |  Diethylene glycol Diethylene glycol#3: Chemical |  Imidazole Imidazole#4: Chemical |  Acetate Acetate#5: Chemical | ChemComp-CL / |  Chloride Chloride#6: Water | ChemComp-HOH / |  Water Water |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 4.29 Å3/Da / Density % sol: 71.33 % |
|---|---|
Crystal grow | Temperature: 293 K / Method: vapor diffusion, hanging drop / pH: 6.5 Details: 0.1M Imidazole pH=6.5 0.9M Sodium acetate, additive 2.0M NDSB-221, VAPOR DIFFUSION, HANGING DROP, temperature 293K |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 19-BM / Wavelength: 0.97923 Å / Beamline: 19-BM / Wavelength: 0.97923 Å |
| Detector | Type: ADSC QUANTUM 210r / Detector: CCD / Date: Jul 9, 2010 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 0.97923 Å / Relative weight: 1 : 0.97923 Å / Relative weight: 1 |
| Reflection | Resolution: 2.26→50 Å / Num. obs: 11234 / % possible obs: 99.8 % / Observed criterion σ(F): 0 / Observed criterion σ(I): -3 / Redundancy: 14 % / Rmerge(I) obs: 0.088 / Net I/σ(I): 36.5 |
| Reflection shell | Resolution: 2.26→2.3 Å / Redundancy: 12.5 % / Mean I/σ(I) obs: 2.16 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1G96 Resolution: 2.26→27.48 Å / Cor.coef. Fo:Fc: 0.955 / Cor.coef. Fo:Fc free: 0.938 / SU B: 9.08 / SU ML: 0.104 / Cross valid method: THROUGHOUT / ESU R Free: 0.16 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.4 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 46.279 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.26→27.48 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.26→2.322 Å / Total num. of bins used: 20
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller















 PDBj
PDBj









