+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3gsl | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of PSD-95 tandem PDZ domains 1 and 2 | ||||||
 Components Components | Disks large homolog 4 | ||||||
 Keywords Keywords |  STRUCTURAL PROTEIN / STRUCTURAL PROTEIN /  PDZ domain / PDZ domain /  tandem / tandem /  PSD-95 / PSD-95 /  DLG4 / SAP-90 / GluR6 / DLG4 / SAP-90 / GluR6 /  Cell junction / Cell junction /  Cell membrane / Cell membrane /  Lipoprotein / Lipoprotein /  Membrane / Membrane /  Palmitate / Palmitate /  Phosphoprotein / Postsynaptic cell membrane / Phosphoprotein / Postsynaptic cell membrane /  SH3 domain / SH3 domain /  Synapse Synapse | ||||||
| Function / homology |  Function and homology information Function and homology informationRHO GTPases activate CIT / positive regulation of AMPA glutamate receptor clustering / neuronal ion channel clustering / P2Y1 nucleotide receptor binding / Neurexins and neuroligins /  beta-1 adrenergic receptor binding / neuroligin family protein binding / structural constituent of postsynaptic density / proximal dendrite / receptor localization to synapse ...RHO GTPases activate CIT / positive regulation of AMPA glutamate receptor clustering / neuronal ion channel clustering / P2Y1 nucleotide receptor binding / Neurexins and neuroligins / beta-1 adrenergic receptor binding / neuroligin family protein binding / structural constituent of postsynaptic density / proximal dendrite / receptor localization to synapse ...RHO GTPases activate CIT / positive regulation of AMPA glutamate receptor clustering / neuronal ion channel clustering / P2Y1 nucleotide receptor binding / Neurexins and neuroligins /  beta-1 adrenergic receptor binding / neuroligin family protein binding / structural constituent of postsynaptic density / proximal dendrite / receptor localization to synapse / positive regulation of neuron projection arborization / regulation of grooming behavior / synaptic vesicle maturation / cerebellar mossy fiber / cellular response to potassium ion / protein localization to synapse / vocalization behavior / LGI-ADAM interactions / Trafficking of AMPA receptors / neuron spine / beta-1 adrenergic receptor binding / neuroligin family protein binding / structural constituent of postsynaptic density / proximal dendrite / receptor localization to synapse / positive regulation of neuron projection arborization / regulation of grooming behavior / synaptic vesicle maturation / cerebellar mossy fiber / cellular response to potassium ion / protein localization to synapse / vocalization behavior / LGI-ADAM interactions / Trafficking of AMPA receptors / neuron spine /  dendritic branch / Activation of Ca-permeable Kainate Receptor / AMPA glutamate receptor clustering / juxtaparanode region of axon / establishment or maintenance of epithelial cell apical/basal polarity / dendritic spine morphogenesis / dendritic branch / Activation of Ca-permeable Kainate Receptor / AMPA glutamate receptor clustering / juxtaparanode region of axon / establishment or maintenance of epithelial cell apical/basal polarity / dendritic spine morphogenesis /  frizzled binding / negative regulation of receptor internalization / postsynaptic neurotransmitter receptor diffusion trapping / neuron projection terminus / dendritic spine organization / frizzled binding / negative regulation of receptor internalization / postsynaptic neurotransmitter receptor diffusion trapping / neuron projection terminus / dendritic spine organization /  acetylcholine receptor binding / positive regulation of synapse assembly / RAF/MAP kinase cascade / Synaptic adhesion-like molecules / neurotransmitter receptor localization to postsynaptic specialization membrane / positive regulation of dendrite morphogenesis / acetylcholine receptor binding / positive regulation of synapse assembly / RAF/MAP kinase cascade / Synaptic adhesion-like molecules / neurotransmitter receptor localization to postsynaptic specialization membrane / positive regulation of dendrite morphogenesis /  beta-2 adrenergic receptor binding / cortical cytoskeleton / regulation of neuronal synaptic plasticity / locomotory exploration behavior / beta-2 adrenergic receptor binding / cortical cytoskeleton / regulation of neuronal synaptic plasticity / locomotory exploration behavior /  regulation of NMDA receptor activity / regulation of NMDA receptor activity /  social behavior / positive regulation of excitatory postsynaptic potential / social behavior / positive regulation of excitatory postsynaptic potential /  kinesin binding / AMPA glutamate receptor complex / neuromuscular process controlling balance / kinesin binding / AMPA glutamate receptor complex / neuromuscular process controlling balance /  excitatory synapse / excitatory synapse /  D1 dopamine receptor binding / Unblocking of NMDA receptors, glutamate binding and activation / D1 dopamine receptor binding / Unblocking of NMDA receptors, glutamate binding and activation /  glutamate receptor binding / positive regulation of protein tyrosine kinase activity / positive regulation of synaptic transmission / glutamate receptor binding / positive regulation of protein tyrosine kinase activity / positive regulation of synaptic transmission /  ionotropic glutamate receptor binding / ionotropic glutamate receptor binding /  extrinsic component of cytoplasmic side of plasma membrane / dendrite cytoplasm / extrinsic component of cytoplasmic side of plasma membrane / dendrite cytoplasm /  synaptic membrane / synaptic membrane /  PDZ domain binding / cell periphery / postsynaptic density membrane / regulation of long-term neuronal synaptic plasticity / PDZ domain binding / cell periphery / postsynaptic density membrane / regulation of long-term neuronal synaptic plasticity /  neuromuscular junction / establishment of protein localization / neuromuscular junction / establishment of protein localization /  cell-cell adhesion / cerebral cortex development / cell-cell adhesion / cerebral cortex development /  kinase binding / cell-cell junction / kinase binding / cell-cell junction /  synaptic vesicle / synaptic vesicle /  cell junction / positive regulation of cytosolic calcium ion concentration / chemical synaptic transmission / postsynapse / cell junction / positive regulation of cytosolic calcium ion concentration / chemical synaptic transmission / postsynapse /  scaffold protein binding / scaffold protein binding /  postsynaptic membrane / basolateral plasma membrane / postsynaptic membrane / basolateral plasma membrane /  protein phosphatase binding / protein-containing complex assembly / protein phosphatase binding / protein-containing complex assembly /  dendritic spine / dendritic spine /  postsynaptic density / neuron projection / postsynaptic density / neuron projection /  signaling receptor binding / signaling receptor binding /  dendrite / dendrite /  synapse / glutamatergic synapse / protein-containing complex binding / synapse / glutamatergic synapse / protein-containing complex binding /  protein kinase binding / protein kinase binding /  endoplasmic reticulum / endoplasmic reticulum /  membrane / membrane /  plasma membrane / plasma membrane /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function | ||||||
| Biological species |   Rattus norvegicus (Norway rat) Rattus norvegicus (Norway rat) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.05 Å MOLECULAR REPLACEMENT / Resolution: 2.05 Å | ||||||
 Authors Authors | Sainlos, M. / Olivier, N.B. / Imperiali, B. | ||||||
 Citation Citation |  Journal: Nat.Chem.Biol. / Year: 2011 Journal: Nat.Chem.Biol. / Year: 2011Title: Biomimetic divalent ligands for the acute disruption of synaptic AMPAR stabilization. Authors: Sainlos, M. / Tigaret, C. / Poujol, C. / Olivier, N.B. / Bard, L. / Breillat, C. / Thiolon, K. / Choquet, D. / Imperiali, B. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3gsl.cif.gz 3gsl.cif.gz | 90.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3gsl.ent.gz pdb3gsl.ent.gz | 68.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3gsl.json.gz 3gsl.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/gs/3gsl https://data.pdbj.org/pub/pdb/validation_reports/gs/3gsl ftp://data.pdbj.org/pub/pdb/validation_reports/gs/3gsl ftp://data.pdbj.org/pub/pdb/validation_reports/gs/3gsl | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 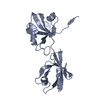
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
| ||||||||
| Details | AUTHORS STATE THAT ONE PROTOMER OF THE ASYMMETRIC UNIT IS THE BIOLOGICAL ASSEMBLY. |
- Components
Components
| #1: Protein | Mass: 20790.654 Da / Num. of mol.: 2 / Fragment: PDZ domains 1 and 2: UNP residues 61-249 Source method: isolated from a genetically manipulated source Details: N-terminal, TEV cleavable octahistidine tag. Three N-terminal residues S-G-S remain after proteolysis. Source: (gene. exp.)   Rattus norvegicus (Norway rat) / Gene: Dlg4, Dlgh4, PSD-95, Psd95 / Plasmid: pET-NO / Production host: Rattus norvegicus (Norway rat) / Gene: Dlg4, Dlgh4, PSD-95, Psd95 / Plasmid: pET-NO / Production host:   Escherichia coli (E. coli) / Strain (production host): BL21 / References: UniProt: P31016 Escherichia coli (E. coli) / Strain (production host): BL21 / References: UniProt: P31016#2: Water | ChemComp-HOH / |  Water WaterSequence details | PDZ DOMAIN 1 LIGAND (ETMA) IS FUSED TO THE C-TERMINUS OF DOMAIN 2. | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.48 Å3/Da / Density % sol: 50.36 % Description: THE ENTRY HAS BEEN UPDATED WITH THE COORDINATES OBTAINED FROM REFINEMENT USING DATA COLLECTED ON 2009-06-26 |
|---|---|
Crystal grow | Temperature: 277 K / Method: vapor diffusion, sitting drop / pH: 7 Details: 35% v/v 2-methyl-2,4-pentanediol, 0.1M Tris-HCl pH 7.0. 0.2M NaCl, VAPOR DIFFUSION, SITTING DROP, temperature 277K |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  NSLS NSLS  / Beamline: X6A / Wavelength: 0.97 Å / Beamline: X6A / Wavelength: 0.97 Å |
| Detector | Type: ADSC QUANTUM 270 / Detector: CCD / Date: Feb 6, 2009 / Details: Toroidal focusing mirror |
| Radiation | Monochromator: Si(111) channel cut / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 0.97 Å / Relative weight: 1 : 0.97 Å / Relative weight: 1 |
| Reflection | Resolution: 2.05→50 Å / Num. all: 26112 / Num. obs: 25616 / % possible obs: 98.1 % / Redundancy: 3.7 % / Biso Wilson estimate: 35.16 Å2 / Rmerge(I) obs: 0.058 / Net I/σ(I): 21.2 |
| Reflection shell | Resolution: 2.05→2.12 Å / Redundancy: 3.6 % / Rmerge(I) obs: 0.349 / Mean I/σ(I) obs: 3.1 / Num. unique all: 2594 / % possible all: 96.3 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB entries 2I1N, 2FE5 Resolution: 2.05→32.31 Å / Cor.coef. Fo:Fc: 0.949 / Cor.coef. Fo:Fc free: 0.909 / Cross valid method: THROUGHOUT / ESU R: 0.251 / ESU R Free: 0.222 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 37.914 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.05→32.31 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.05→2.103 Å / Total num. of bins used: 20
|
 Movie
Movie Controller
Controller













 PDBj
PDBj




