+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3fd7 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
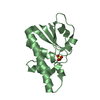
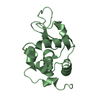
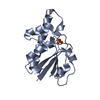
| Title | Crystal structure of Onconase C87A/C104A-ONC | |||||||||
 Components Components | Protein P-30 | |||||||||
 Keywords Keywords |  HYDROLASE / HYDROLASE /  Onconase / C-terminal disulfide bond / Onconase / C-terminal disulfide bond /  Endonuclease / Endonuclease /  Nuclease / Nuclease /  Pyrrolidone carboxylic acid Pyrrolidone carboxylic acid | |||||||||
| Function / homology |  Function and homology information Function and homology information Hydrolases; Acting on ester bonds; Endoribonucleases producing 3'-phosphomonoesters / Hydrolases; Acting on ester bonds; Endoribonucleases producing 3'-phosphomonoesters /  endonuclease activity / endonuclease activity /  nucleic acid binding nucleic acid bindingSimilarity search - Function | |||||||||
| Biological species |  Rana pipiens (northern leopard frog) Rana pipiens (northern leopard frog) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 1.531 Å molecular replacement / Resolution: 1.531 Å | |||||||||
 Authors Authors | Neumann, P. / Schulenburg, C. / Arnold, U. / Ulbrich-Hofmann, R. / Stubbs, M.T. | |||||||||
 Citation Citation |  Journal: Chembiochem / Year: 2010 Journal: Chembiochem / Year: 2010Title: Impact of the C-terminal disulfide bond on the folding and stability of onconase. Authors: Schulenburg, C. / Weininger, U. / Neumann, P. / Meiselbach, H. / Stubbs, M.T. / Sticht, H. / Balbach, J. / Ulbrich-Hofmann, R. / Arnold, U. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3fd7.cif.gz 3fd7.cif.gz | 118.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3fd7.ent.gz pdb3fd7.ent.gz | 90.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3fd7.json.gz 3fd7.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/fd/3fd7 https://data.pdbj.org/pub/pdb/validation_reports/fd/3fd7 ftp://data.pdbj.org/pub/pdb/validation_reports/fd/3fd7 ftp://data.pdbj.org/pub/pdb/validation_reports/fd/3fd7 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  2kb6C  1oncS C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| 3 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 11781.518 Da / Num. of mol.: 2 / Mutation: C87A, C104A Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Rana pipiens (northern leopard frog) / Plasmid: pET26b(+) / Production host: Rana pipiens (northern leopard frog) / Plasmid: pET26b(+) / Production host:   Escherichia coli BL21(DE3) (bacteria) / Strain (production host): BL21(DE3) Escherichia coli BL21(DE3) (bacteria) / Strain (production host): BL21(DE3)References: UniProt: P22069,  Hydrolases; Acting on ester bonds; Endoribonucleases producing 3'-phosphomonoesters Hydrolases; Acting on ester bonds; Endoribonucleases producing 3'-phosphomonoesters#2: Chemical | ChemComp-SO4 /  Sulfate Sulfate#3: Chemical |  Glycerol Glycerol#4: Chemical | ChemComp-EDO / |  Ethylene glycol Ethylene glycol#5: Water | ChemComp-HOH / |  Water Water |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.17 Å3/Da / Density % sol: 43.19 % |
|---|---|
Crystal grow | Temperature: 293 K / Method: vapor diffusion, hanging drop / pH: 5.5 Details: 0.2M ammoniumsulfate, 30% PEG 4000, pH 5.5, VAPOR DIFFUSION, HANGING DROP, temperature 293K |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU MICROMAX-007 / Wavelength: 1.5418 Å ROTATING ANODE / Type: RIGAKU MICROMAX-007 / Wavelength: 1.5418 Å |
| Detector | Type: RIGAKU RAXIS IV++ / Detector: IMAGE PLATE / Date: Mar 12, 2008 / Details: GRAPHITE |
| Radiation | Monochromator: GRAPHITE / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 1.5418 Å / Relative weight: 1 : 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 1.53→26.864 Å / Num. all: 31616 / Num. obs: 31390 / % possible obs: 99.3 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Redundancy: 23.35 % / Biso Wilson estimate: 21.64 Å2 / Rmerge(I) obs: 0.073 / Rsym value: 0.074 / Net I/σ(I): 32.52 |
| Reflection shell | Resolution: 1.53→1.62 Å / Redundancy: 18.78 % / Rmerge(I) obs: 0.557 / Mean I/σ(I) obs: 5.76 / Num. unique all: 4680 / Rsym value: 0.572 / % possible all: 95.5 |
-Phasing
Phasing | Method:  molecular replacement molecular replacement | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Phasing MR | Rfactor: 47.42 / Packing: 0
|
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 1ONC Resolution: 1.531→26.864 Å / Occupancy max: 1 / Occupancy min: 0.3 / FOM work R set: 0.898 / SU ML: 0.18 / Cross valid method: THROUGHOUT / σ(F): 1.37 / σ(I): 0 / Stereochemistry target values: ML
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL / Bsol: 50.834 Å2 / ksol: 0.372 e/Å3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 55.25 Å2 / Biso mean: 18.07 Å2 / Biso min: 7.85 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.531→26.864 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Total num. of bins used: 11
|
 Movie
Movie Controller
Controller










 PDBj
PDBj








