+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3dv4 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of SAG506-01, tetragonal, crystal 1 | ||||||
 Components Components |
| ||||||
 Keywords Keywords |  IMMUNE SYSTEM / IMMUNE SYSTEM /  antibody / KDO / twinning / pseudo-symmetry antibody / KDO / twinning / pseudo-symmetry | ||||||
| Function / homology |  Function and homology information Function and homology information | ||||||
| Biological species |   Mus Musculus (house mouse) Mus Musculus (house mouse) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 1.95 Å molecular replacement / Resolution: 1.95 Å | ||||||
 Authors Authors | Brooks, C.L. / Blackler, R.J. / Gerstenbruch, S. / Kosma, P. / Muller-Loennies, S. / Brade, H. / Evans, S.V. | ||||||
 Citation Citation |  Journal: Acta Crystallogr.,Sect.D / Year: 2008 Journal: Acta Crystallogr.,Sect.D / Year: 2008Title: Pseudo-symmetry and twinning in crystals of homologous antibody Fv fragments. Authors: Brooks, C.L. / Blackler, R.J. / Gerstenbruch, S. / Kosma, P. / Muller-Loennies, S. / Brade, H. / Evans, S.V. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3dv4.cif.gz 3dv4.cif.gz | 57.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3dv4.ent.gz pdb3dv4.ent.gz | 44.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3dv4.json.gz 3dv4.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/dv/3dv4 https://data.pdbj.org/pub/pdb/validation_reports/dv/3dv4 ftp://data.pdbj.org/pub/pdb/validation_reports/dv/3dv4 ftp://data.pdbj.org/pub/pdb/validation_reports/dv/3dv4 | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Components on special symmetry positions |
|
- Components
Components
-Antibody , 2 types, 2 molecules AB
| #1: Antibody | Mass: 12353.851 Da / Num. of mol.: 1 / Fragment: variable region fragment (Fv) Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Mus Musculus (house mouse) / Plasmid: pSFJ8 / Production host: Mus Musculus (house mouse) / Plasmid: pSFJ8 / Production host:   Escherichia coli (E. coli) / Strain (production host): TG1 / References: UniProt: A0N262*PLUS Escherichia coli (E. coli) / Strain (production host): TG1 / References: UniProt: A0N262*PLUS |
|---|---|
| #2: Antibody | Mass: 13442.986 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Mus Musculus (house mouse) / Production host: Mus Musculus (house mouse) / Production host:   Escherichia coli (E. coli) / References: UniProt: A2NU21*PLUS Escherichia coli (E. coli) / References: UniProt: A2NU21*PLUS |
-Sugars , 1 types, 1 molecules 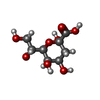
| #4: Sugar | ChemComp-KDO / |
|---|
-Non-polymers , 3 types, 133 molecules 




| #3: Chemical | ChemComp-MG / |
|---|---|
| #5: Chemical | ChemComp-PG4 /  Polyethylene glycol Polyethylene glycol |
| #6: Water | ChemComp-HOH /  Water Water |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.19 Å3/Da / Density % sol: 43.8 % |
|---|---|
Crystal grow | Temperature: 298 K / Method: vapor diffusion / pH: 8.5 Details: PEG 4000, MgCl2, pH 8.5, vapor diffusion, temperature 298K |
-Data collection
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU MICROMAX-002 / Wavelength: 1.5418 Å ROTATING ANODE / Type: RIGAKU MICROMAX-002 / Wavelength: 1.5418 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Type: RIGAKU RAXIS IV++ / Detector: IMAGE PLATE | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength : 1.5418 Å / Relative weight: 1 : 1.5418 Å / Relative weight: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection | Resolution: 1.95→19.81 Å / Num. obs: 17255 / % possible obs: 99.9 % / Redundancy: 9.4 % / Rmerge(I) obs: 0.07 / Χ2: 1 / Net I/σ(I): 14.7 / Scaling rejects: 1226 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection shell |
|
-Phasing
Phasing | Method:  molecular replacement molecular replacement | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Phasing MR | Rfactor: 44.54 / Model details: Phaser MODE: MR_AUTO
|
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT / Resolution: 1.95→19.81 Å / Occupancy max: 1 / Occupancy min: 1 / FOM work R set: 0.68 / SU ML: 0.44 / σ(F): 1.4 / Stereochemistry target values: ML MOLECULAR REPLACEMENT / Resolution: 1.95→19.81 Å / Occupancy max: 1 / Occupancy min: 1 / FOM work R set: 0.68 / SU ML: 0.44 / σ(F): 1.4 / Stereochemistry target values: ML
| |||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL / Bsol: 53.158 Å2 / ksol: 0.319 e/Å3 | |||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 66.35 Å2 / Biso mean: 37.986 Å2 / Biso min: 19.22 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.95→19.81 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Total num. of bins used: 6
|
 Movie
Movie Controller
Controller















 PDBj
PDBj




