[English] 日本語
 Yorodumi
Yorodumi- PDB-3d1i: Structure of the Thioalkalivibrio nitratireducens cytochrome c ni... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3d1i | ||||||
|---|---|---|---|---|---|---|---|
| Title | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase in a complex with nitrite | ||||||
 Components Components | Eight-heme nitrite reductase | ||||||
 Keywords Keywords |  OXIDOREDUCTASE / OXIDOREDUCTASE /  cytochrome C nitrite reductase / NRFA / cytochrome C nitrite reductase / NRFA /  Sulfite reductase Sulfite reductase | ||||||
| Function / homology |  Function and homology information Function and homology informationammonium ion metabolic process /  nitrite reductase (cytochrome; ammonia-forming) / nitrite reductase (cytochrome, ammonia-forming) activity / anaerobic electron transport chain / nitrate assimilation / outer membrane-bounded periplasmic space / nitrite reductase (cytochrome; ammonia-forming) / nitrite reductase (cytochrome, ammonia-forming) activity / anaerobic electron transport chain / nitrate assimilation / outer membrane-bounded periplasmic space /  periplasmic space / periplasmic space /  calcium ion binding / calcium ion binding /  heme binding heme bindingSimilarity search - Function | ||||||
| Biological species |  Thioalkalivibrio nitratireducens (bacteria) Thioalkalivibrio nitratireducens (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.8 Å MOLECULAR REPLACEMENT / Resolution: 1.8 Å | ||||||
 Authors Authors | Polyakov, K.M. / Boyko, K.M. / Slutsky, A. / Tikhonova, T.V. / Antipov, A.N. / Zvyagilskaya, R.A. / Popov, A.N. / Lamzin, V.S. / Bourenkov, G.P. / Popov, V.O. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 2009 Journal: J.Mol.Biol. / Year: 2009Title: High-resolution structural analysis of a novel octaheme cytochrome c nitrite reductase from the haloalkaliphilic bacterium Thioalkalivibrio nitratireducens Authors: Polyakov, K.M. / Boyko, K.M. / Tikhonova, T.V. / Slutsky, A. / Antipov, A.N. / Zvyagilskaya, R.A. / Popov, A.N. / Bourenkov, G.P. / Lamzin, V.S. / Popov, V.O. #1: Journal: Acta Crystallogr.,Sect.F / Year: 2006 Title: Crystallization and preliminary X-ray analysis of cytochrome c nitrite reductase from Thioalkalivibrio nitratireducens Authors: Boyko, K.M. / Polyakov, K.M. / Tikhonova, T.V. / Slutsky, A. / Antipov, A.N. / Zvyagilskaya, R.A. / Bourenkov, G.P. / Popov, A.N. / Lamzin, V.S. / Popov, V.O. #2: Journal: Biochim.Biophys.Acta / Year: 2006 Title: Molecular and catalytic properties of a novel cytochrome c nitrite reductase from nitrate-reducing haloalkaliphilic sulfur-oxidizing bacterium Thioalkalivibrio nitratireducens Authors: Tikhonova, T.V. / Slutsky, A. / Antipov, A.N. / Boyko, K.M. / Polyakov, K.M. / Sorokin, D.Y. / Zvyagilskaya, R.A. / Popov, V.O. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3d1i.cif.gz 3d1i.cif.gz | 445.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3d1i.ent.gz pdb3d1i.ent.gz | 370.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3d1i.json.gz 3d1i.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/d1/3d1i https://data.pdbj.org/pub/pdb/validation_reports/d1/3d1i ftp://data.pdbj.org/pub/pdb/validation_reports/d1/3d1i ftp://data.pdbj.org/pub/pdb/validation_reports/d1/3d1i | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  2ot4SC  2zo5C S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||||
| Unit cell |
| ||||||||||||
| Components on special symmetry positions |
|
- Components
Components
-Protein , 1 types, 2 molecules AB
| #1: Protein | Mass: 59491.309 Da / Num. of mol.: 2 / Source method: isolated from a natural source Source: (natural)  Thioalkalivibrio nitratireducens (bacteria) Thioalkalivibrio nitratireducens (bacteria)Strain: ALEN 2 / References: UniProt: Q5F2I3, UniProt: L0DSL2*PLUS |
|---|
-Non-polymers , 7 types, 1162 molecules 

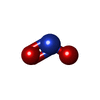


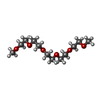







| #2: Chemical | ChemComp-HEC /  Heme C Heme C#3: Chemical | ChemComp-CA / #4: Chemical |  Nitrite Nitrite#5: Chemical | #6: Chemical | ChemComp-PG4 /  Polyethylene glycol Polyethylene glycol#7: Chemical |  Polyethylene glycol Polyethylene glycol#8: Water | ChemComp-HOH / |  Water Water |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 5.07 Å3/Da / Density % sol: 75.73 % |
|---|---|
Crystal grow | Temperature: 278 K / Method: vapor diffusion, hanging drop / pH: 8.5 Details: The drop contained 5ml of enzyme solution (14.5mg/ml) in 0.005M Tris-borat buffer (PH8.7) and 5ml reservoir solution. The reservoir solution contained 27% PEG400, 0.18M sodium citrate in 0. ...Details: The drop contained 5ml of enzyme solution (14.5mg/ml) in 0.005M Tris-borat buffer (PH8.7) and 5ml reservoir solution. The reservoir solution contained 27% PEG400, 0.18M sodium citrate in 0.09M Tris buffer (PH8.5), complex with nitrite ion was obtained by soaking crystal in reservoir solution with 100mM sodium nitrite by 30 minute, VAPOR DIFFUSION, HANGING DROP, temperature 278K |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  EMBL/DESY, HAMBURG EMBL/DESY, HAMBURG  / Beamline: X13 / Wavelength: 0.801 / Beamline: X13 / Wavelength: 0.801 |
| Detector | Type: MAR CCD 165 mm / Detector: CCD |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 0.801 Å / Relative weight: 1 : 0.801 Å / Relative weight: 1 |
| Reflection | Resolution: 1.8→13 Å / Num. obs: 228600 / % possible obs: 99.7 % / Rmerge(I) obs: 0.079 / Net I/σ(I): 16 |
| Reflection shell | Resolution: 1.8→1.82 Å / Rmerge(I) obs: 0.6 / % possible all: 98.8 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 2OT4 Resolution: 1.8→11.99 Å / Cor.coef. Fo:Fc: 0.967 / Cor.coef. Fo:Fc free: 0.962 / SU B: 1.371 / SU ML: 0.043 / Cross valid method: THROUGHOUT / ESU R: 0.067 / ESU R Free: 0.067 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: BABINET MODEL WITH MASK | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 16.63 Å2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.155 Å / Luzzati sigma a obs: 0.029 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.8→11.99 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.8→1.847 Å / Total num. of bins used: 20
|
 Movie
Movie Controller
Controller












 PDBj
PDBj













