[English] 日本語
 Yorodumi
Yorodumi- PDB-2vt8: Structure of a conserved dimerisation domain within Fbox7 and PI31 -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2vt8 | ||||||
|---|---|---|---|---|---|---|---|
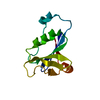
| Title | Structure of a conserved dimerisation domain within Fbox7 and PI31 | ||||||
 Components Components | PROTEASOME INHIBITOR PI31 SUBUNIT | ||||||
 Keywords Keywords | HYDROLASE INHIBITOR / POLYMORPHISM | ||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of proteasomal protein catabolic process / Regulation of ornithine decarboxylase (ODC) / proteasome core complex / Cross-presentation of soluble exogenous antigens (endosomes) /  Somitogenesis / Somitogenesis /  endopeptidase inhibitor activity / endopeptidase inhibitor activity /  proteasome binding / Regulation of activated PAK-2p34 by proteasome mediated degradation / Autodegradation of Cdh1 by Cdh1:APC/C / APC/C:Cdc20 mediated degradation of Securin ...negative regulation of proteasomal protein catabolic process / Regulation of ornithine decarboxylase (ODC) / proteasome core complex / Cross-presentation of soluble exogenous antigens (endosomes) / proteasome binding / Regulation of activated PAK-2p34 by proteasome mediated degradation / Autodegradation of Cdh1 by Cdh1:APC/C / APC/C:Cdc20 mediated degradation of Securin ...negative regulation of proteasomal protein catabolic process / Regulation of ornithine decarboxylase (ODC) / proteasome core complex / Cross-presentation of soluble exogenous antigens (endosomes) /  Somitogenesis / Somitogenesis /  endopeptidase inhibitor activity / endopeptidase inhibitor activity /  proteasome binding / Regulation of activated PAK-2p34 by proteasome mediated degradation / Autodegradation of Cdh1 by Cdh1:APC/C / APC/C:Cdc20 mediated degradation of Securin / Asymmetric localization of PCP proteins / SCF-beta-TrCP mediated degradation of Emi1 / NIK-->noncanonical NF-kB signaling / Ubiquitin-dependent degradation of Cyclin D / AUF1 (hnRNP D0) binds and destabilizes mRNA / TNFR2 non-canonical NF-kB pathway / Assembly of the pre-replicative complex / Vpu mediated degradation of CD4 / Degradation of DVL / Ubiquitin Mediated Degradation of Phosphorylated Cdc25A / Dectin-1 mediated noncanonical NF-kB signaling / Hh mutants are degraded by ERAD / Cdc20:Phospho-APC/C mediated degradation of Cyclin A / Degradation of AXIN / Defective CFTR causes cystic fibrosis / Degradation of GLI1 by the proteasome / Hedgehog ligand biogenesis / Activation of NF-kappaB in B cells / Negative regulation of NOTCH4 signaling / GSK3B and BTRC:CUL1-mediated-degradation of NFE2L2 / G2/M Checkpoints / Vif-mediated degradation of APOBEC3G / Autodegradation of the E3 ubiquitin ligase COP1 / Hedgehog 'on' state / Regulation of RUNX3 expression and activity / Degradation of GLI2 by the proteasome / GLI3 is processed to GLI3R by the proteasome / MAPK6/MAPK4 signaling / FBXL7 down-regulates AURKA during mitotic entry and in early mitosis / APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 / ABC-family proteins mediated transport / Degradation of beta-catenin by the destruction complex / Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha / CDK-mediated phosphorylation and removal of Cdc6 / CLEC7A (Dectin-1) signaling / SCF(Skp2)-mediated degradation of p27/p21 / Regulation of expression of SLITs and ROBOs / FCERI mediated NF-kB activation / Regulation of PTEN stability and activity / Interleukin-1 signaling / Orc1 removal from chromatin / Regulation of RAS by GAPs / Separation of Sister Chromatids / Regulation of RUNX2 expression and activity / The role of GTSE1 in G2/M progression after G2 checkpoint / UCH proteinases / KEAP1-NFE2L2 pathway / Antigen processing: Ubiquitination & Proteasome degradation / Downstream TCR signaling / proteasome binding / Regulation of activated PAK-2p34 by proteasome mediated degradation / Autodegradation of Cdh1 by Cdh1:APC/C / APC/C:Cdc20 mediated degradation of Securin / Asymmetric localization of PCP proteins / SCF-beta-TrCP mediated degradation of Emi1 / NIK-->noncanonical NF-kB signaling / Ubiquitin-dependent degradation of Cyclin D / AUF1 (hnRNP D0) binds and destabilizes mRNA / TNFR2 non-canonical NF-kB pathway / Assembly of the pre-replicative complex / Vpu mediated degradation of CD4 / Degradation of DVL / Ubiquitin Mediated Degradation of Phosphorylated Cdc25A / Dectin-1 mediated noncanonical NF-kB signaling / Hh mutants are degraded by ERAD / Cdc20:Phospho-APC/C mediated degradation of Cyclin A / Degradation of AXIN / Defective CFTR causes cystic fibrosis / Degradation of GLI1 by the proteasome / Hedgehog ligand biogenesis / Activation of NF-kappaB in B cells / Negative regulation of NOTCH4 signaling / GSK3B and BTRC:CUL1-mediated-degradation of NFE2L2 / G2/M Checkpoints / Vif-mediated degradation of APOBEC3G / Autodegradation of the E3 ubiquitin ligase COP1 / Hedgehog 'on' state / Regulation of RUNX3 expression and activity / Degradation of GLI2 by the proteasome / GLI3 is processed to GLI3R by the proteasome / MAPK6/MAPK4 signaling / FBXL7 down-regulates AURKA during mitotic entry and in early mitosis / APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 / ABC-family proteins mediated transport / Degradation of beta-catenin by the destruction complex / Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha / CDK-mediated phosphorylation and removal of Cdc6 / CLEC7A (Dectin-1) signaling / SCF(Skp2)-mediated degradation of p27/p21 / Regulation of expression of SLITs and ROBOs / FCERI mediated NF-kB activation / Regulation of PTEN stability and activity / Interleukin-1 signaling / Orc1 removal from chromatin / Regulation of RAS by GAPs / Separation of Sister Chromatids / Regulation of RUNX2 expression and activity / The role of GTSE1 in G2/M progression after G2 checkpoint / UCH proteinases / KEAP1-NFE2L2 pathway / Antigen processing: Ubiquitination & Proteasome degradation / Downstream TCR signaling /  Neddylation / RUNX1 regulates transcription of genes involved in differentiation of HSCs / ER-Phagosome pathway / ubiquitin-dependent protein catabolic process / proteasome-mediated ubiquitin-dependent protein catabolic process / Ub-specific processing proteases / protein heterodimerization activity / perinuclear region of cytoplasm / Neddylation / RUNX1 regulates transcription of genes involved in differentiation of HSCs / ER-Phagosome pathway / ubiquitin-dependent protein catabolic process / proteasome-mediated ubiquitin-dependent protein catabolic process / Ub-specific processing proteases / protein heterodimerization activity / perinuclear region of cytoplasm /  endoplasmic reticulum / protein homodimerization activity / endoplasmic reticulum / protein homodimerization activity /  nucleoplasm / nucleoplasm /  membrane / membrane /  cytosol cytosolSimilarity search - Function | ||||||
| Biological species |   HOMO SAPIENS (human) HOMO SAPIENS (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 2.6 Å MOLECULAR REPLACEMENT / Resolution: 2.6 Å | ||||||
 Authors Authors | Kirk, R.J. / Murray-Rust, J. / Knowles, P.P. / Laman, H. / McDonald, N.Q. | ||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 2008 Journal: J.Biol.Chem. / Year: 2008Title: Structure of a Conserved Dimerization Domain within the F-Box Protein Fbxo7 and the Pi31 Proteasome Inhibitor. Authors: Kirk, R.J. / Laman, H. / Knowles, P.P. / Murray-Rust, J. / Lomonosov, M. / Meziane, E.K. / McDonald, N.Q. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2vt8.cif.gz 2vt8.cif.gz | 66.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2vt8.ent.gz pdb2vt8.ent.gz | 49.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2vt8.json.gz 2vt8.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/vt/2vt8 https://data.pdbj.org/pub/pdb/validation_reports/vt/2vt8 ftp://data.pdbj.org/pub/pdb/validation_reports/vt/2vt8 ftp://data.pdbj.org/pub/pdb/validation_reports/vt/2vt8 | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
| ||||||||
| Noncrystallographic symmetry (NCS) | NCS oper: (Code: given Matrix: (-0.9756, 0.03852, 0.2162), Vector  : : |
- Components
Components
| #1: Protein | Mass: 17198.361 Da / Num. of mol.: 2 / Fragment: RESIDUES 1-151 / Mutation: YES Source method: isolated from a genetically manipulated source Source: (gene. exp.)   HOMO SAPIENS (human) / Production host: HOMO SAPIENS (human) / Production host:   ESCHERICHIA COLI (E. coli) / Strain (production host): BL21(DE3) / References: UniProt: Q92530 ESCHERICHIA COLI (E. coli) / Strain (production host): BL21(DE3) / References: UniProt: Q92530#2: Water | ChemComp-HOH / |  Water WaterCompound details | ENGINEERED | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.28 Å3/Da / Density % sol: 46 % / Description: NONE |
|---|---|
Crystal grow | pH: 8 / Details: 20% PEG 3350, 0.1 M AMMONIUM IODIDE, pH 8 |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU RU200 / Wavelength: 1.5418 ROTATING ANODE / Type: RIGAKU RU200 / Wavelength: 1.5418 |
| Detector | Type: MARRESEARCH / Detector: IMAGE PLATE / Date: Jan 6, 2004 / Details: MIRRORS |
| Radiation | Monochromator: YALE MIRRORS / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 1.5418 Å / Relative weight: 1 : 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 2.6→28.1 Å / Num. obs: 9607 / % possible obs: 99.6 % / Observed criterion σ(I): 2 / Redundancy: 7.1 % / Rmerge(I) obs: 0.08 / Net I/σ(I): 20.1 |
| Reflection shell | Resolution: 2.6→2.74 Å / Redundancy: 6.5 % / Rmerge(I) obs: 0.39 / Mean I/σ(I) obs: 4.1 / % possible all: 97.9 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PART-REFINED MODEL OF A L7M SEMET DERIVATIVE OBTAINED BY SAD PHASING Resolution: 2.6→28.09 Å / Cor.coef. Fo:Fc: 0.941 / Cor.coef. Fo:Fc free: 0.913 / SU B: 11.888 / SU ML: 0.252 / Cross valid method: THROUGHOUT / ESU R: 1.262 / ESU R Free: 0.331 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 42.62 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.6→28.09 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller











 PDBj
PDBj


