[English] 日本語
 Yorodumi
Yorodumi- PDB-2q7r: Crystal structure of human FLAP with an iodinated analog of MK-591 -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2q7r | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of human FLAP with an iodinated analog of MK-591 | ||||||
 Components Components | Arachidonate 5-lipoxygenase-activating protein | ||||||
 Keywords Keywords |  MEMBRANE PROTEIN / LIPID TRANSPORT / FLAP / MEMBRANE PROTEIN / LIPID TRANSPORT / FLAP /  MAPEG MAPEG | ||||||
| Function / homology |  Function and homology information Function and homology informationleukotriene production involved in inflammatory response /  arachidonate 5-lipoxygenase activity / arachidonate 5-lipoxygenase activity /  leukotriene-C4 synthase activity / Synthesis of Lipoxins (LX) / Synthesis of 5-eicosatetraenoic acids / lipoxygenase pathway / Synthesis of Leukotrienes (LT) and Eoxins (EX) / positive regulation of acute inflammatory response / leukotriene biosynthetic process / leukotriene-C4 synthase activity / Synthesis of Lipoxins (LX) / Synthesis of 5-eicosatetraenoic acids / lipoxygenase pathway / Synthesis of Leukotrienes (LT) and Eoxins (EX) / positive regulation of acute inflammatory response / leukotriene biosynthetic process /  glutathione peroxidase activity ...leukotriene production involved in inflammatory response / glutathione peroxidase activity ...leukotriene production involved in inflammatory response /  arachidonate 5-lipoxygenase activity / arachidonate 5-lipoxygenase activity /  leukotriene-C4 synthase activity / Synthesis of Lipoxins (LX) / Synthesis of 5-eicosatetraenoic acids / lipoxygenase pathway / Synthesis of Leukotrienes (LT) and Eoxins (EX) / positive regulation of acute inflammatory response / leukotriene biosynthetic process / leukotriene-C4 synthase activity / Synthesis of Lipoxins (LX) / Synthesis of 5-eicosatetraenoic acids / lipoxygenase pathway / Synthesis of Leukotrienes (LT) and Eoxins (EX) / positive regulation of acute inflammatory response / leukotriene biosynthetic process /  glutathione peroxidase activity / glutathione peroxidase activity /  arachidonic acid binding / protein homotrimerization / arachidonic acid binding / protein homotrimerization /  glutathione transferase activity / glutathione transferase activity /  enzyme activator activity / cellular response to calcium ion / enzyme activator activity / cellular response to calcium ion /  nuclear envelope / nuclear envelope /  nuclear membrane / protein-containing complex binding / endoplasmic reticulum membrane / nuclear membrane / protein-containing complex binding / endoplasmic reticulum membrane /  enzyme binding / enzyme binding /  endoplasmic reticulum / endoplasmic reticulum /  membrane / identical protein binding membrane / identical protein bindingSimilarity search - Function | ||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  SAD / Resolution: 4 Å SAD / Resolution: 4 Å | ||||||
 Authors Authors | Ferguson, A.D. | ||||||
 Citation Citation |  Journal: Science / Year: 2007 Journal: Science / Year: 2007Title: Crystal structure of inhibitor-bound human 5-lipoxygenase-activating protein. Authors: Ferguson, A.D. / McKeever, B.M. / Xu, S. / Wisniewski, D. / Miller, D.K. / Yamin, T.T. / Spencer, R.H. / Chu, L. / Ujjainwalla, F. / Cunningham, B.R. / Evans, J.F. / Becker, J.W. | ||||||
| History |
| ||||||
| Remark 650 | HELIX DETERMINATION METHOD: AUTHOR DETERMINED |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2q7r.cif.gz 2q7r.cif.gz | 182.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2q7r.ent.gz pdb2q7r.ent.gz | 155.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2q7r.json.gz 2q7r.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/q7/2q7r https://data.pdbj.org/pub/pdb/validation_reports/q7/2q7r ftp://data.pdbj.org/pub/pdb/validation_reports/q7/2q7r ftp://data.pdbj.org/pub/pdb/validation_reports/q7/2q7r | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
| ||||||||
| Details | The biological unit is a trimer |
- Components
Components
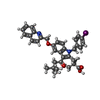
| #1: Protein | Mass: 18252.596 Da / Num. of mol.: 6 / Mutation: K148A Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: ALOX5AP, FLAP / Plasmid: pET28a / Species (production host): Escherichia coli / Production host: Homo sapiens (human) / Gene: ALOX5AP, FLAP / Plasmid: pET28a / Species (production host): Escherichia coli / Production host:   Escherichia coli BL21(DE3) (bacteria) / Strain (production host): BL21(DE3) / References: UniProt: P20292 Escherichia coli BL21(DE3) (bacteria) / Strain (production host): BL21(DE3) / References: UniProt: P20292#2: Chemical | ChemComp-3CS / |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 5.21 Å3/Da / Density % sol: 76.41 % |
|---|---|
Crystal grow | Temperature: 298 K / pH: 5.6 Details: 10% PEG 4000, 0.32 LiCl2, 1 mM TCEP, pH 5.6, VAPOR DIFFUSION, SITTING DROP, temperature 298K, pH 5.60 |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  NSLS NSLS  / Beamline: X25 / Wavelength: 0.9797 / Beamline: X25 / Wavelength: 0.9797 |
| Detector | Type: ADSC QUANTUM 315 / Detector: CCD / Date: Jun 1, 2006 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 0.9797 Å / Relative weight: 1 : 0.9797 Å / Relative weight: 1 |
| Reflection | Resolution: 4→45 Å / Num. obs: 20007 / % possible obs: 99.3 % / Observed criterion σ(I): 0 / Redundancy: 15.5 % / Biso Wilson estimate: 98.721 Å2 / Rmerge(I) obs: 0.111 / Net I/σ(I): 20.6 |
| Reflection shell | Resolution: 4→4.24 Å / Redundancy: 15.5 % / Mean I/σ(I) obs: 3.29 / % possible all: 99.9 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  SAD / Resolution: 4→20 Å / Cross valid method: THROUGHOUT / σ(F): 0 / Stereochemistry target values: Engh & Huber SAD / Resolution: 4→20 Å / Cross valid method: THROUGHOUT / σ(F): 0 / Stereochemistry target values: Engh & Huber
| ||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 120.76 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 4→20 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 4→4.24 Å / Total num. of bins used: 9
|
 Movie
Movie Controller
Controller











 PDBj
PDBj