+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2ms4 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Cyclophilin a complexed with a fragment of crk-ii | ||||||
 Components Components |
| ||||||
 Keywords Keywords |  ISOMERASE / ISOMERASE /  CYCLOPHILIN A CYCLOPHILIN A | ||||||
| Function / homology |  Function and homology information Function and homology informationresponse to hepatocyte growth factor / regulation of intracellular signal transduction / cellular response to endothelin / helper T cell diapedesis / cerebellar neuron development / response to cholecystokinin / postsynaptic specialization assembly / protein phosphorylated amino acid binding / regulation of T cell migration / reelin-mediated signaling pathway ...response to hepatocyte growth factor / regulation of intracellular signal transduction / cellular response to endothelin / helper T cell diapedesis / cerebellar neuron development / response to cholecystokinin / postsynaptic specialization assembly / protein phosphorylated amino acid binding / regulation of T cell migration / reelin-mediated signaling pathway / regulation of dendrite development / positive regulation of skeletal muscle acetylcholine-gated channel clustering / response to yeast / negative regulation of wound healing / negative regulation of natural killer cell mediated cytotoxicity /  regulation of protein binding / negative regulation of cell motility / negative regulation of protein K48-linked ubiquitination / negative regulation of viral life cycle / regulation of apoptotic signaling pathway / cell adhesion molecule production / lipid droplet organization / ARMS-mediated activation / regulation of protein binding / negative regulation of cell motility / negative regulation of protein K48-linked ubiquitination / negative regulation of viral life cycle / regulation of apoptotic signaling pathway / cell adhesion molecule production / lipid droplet organization / ARMS-mediated activation /  heparan sulfate binding / regulation of viral genome replication / MET receptor recycling / protein localization to membrane / leukocyte chemotaxis / negative regulation of stress-activated MAPK cascade / heparan sulfate binding / regulation of viral genome replication / MET receptor recycling / protein localization to membrane / leukocyte chemotaxis / negative regulation of stress-activated MAPK cascade /  endothelial cell activation / MET activates RAP1 and RAC1 / endothelial cell activation / MET activates RAP1 and RAC1 /  virion binding / Basigin interactions / cellular response to insulin-like growth factor stimulus / virion binding / Basigin interactions / cellular response to insulin-like growth factor stimulus /  cyclosporin A binding / establishment of cell polarity / cyclosporin A binding / establishment of cell polarity /  regulation of GTPase activity / p130Cas linkage to MAPK signaling for integrins / Minus-strand DNA synthesis / Plus-strand DNA synthesis / Uncoating of the HIV Virion / positive regulation of smooth muscle cell migration / dendrite development / Early Phase of HIV Life Cycle / positive regulation of Rac protein signal transduction / Integration of provirus / APOBEC3G mediated resistance to HIV-1 infection / regulation of cell adhesion mediated by integrin / Calcineurin activates NFAT / viral release from host cell / cellular response to nitric oxide / regulation of GTPase activity / p130Cas linkage to MAPK signaling for integrins / Minus-strand DNA synthesis / Plus-strand DNA synthesis / Uncoating of the HIV Virion / positive regulation of smooth muscle cell migration / dendrite development / Early Phase of HIV Life Cycle / positive regulation of Rac protein signal transduction / Integration of provirus / APOBEC3G mediated resistance to HIV-1 infection / regulation of cell adhesion mediated by integrin / Calcineurin activates NFAT / viral release from host cell / cellular response to nitric oxide /  regulation of signal transduction / ephrin receptor signaling pathway / Binding and entry of HIV virion / positive regulation of viral genome replication / protein peptidyl-prolyl isomerization / PTK6 Regulates RHO GTPases, RAS GTPase and MAP kinases / signaling adaptor activity / negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway / positive regulation of substrate adhesion-dependent cell spreading / positive regulation of protein dephosphorylation / cellular response to transforming growth factor beta stimulus / regulation of signal transduction / ephrin receptor signaling pathway / Binding and entry of HIV virion / positive regulation of viral genome replication / protein peptidyl-prolyl isomerization / PTK6 Regulates RHO GTPases, RAS GTPase and MAP kinases / signaling adaptor activity / negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway / positive regulation of substrate adhesion-dependent cell spreading / positive regulation of protein dephosphorylation / cellular response to transforming growth factor beta stimulus /  insulin-like growth factor receptor binding / insulin-like growth factor receptor binding /  cytoskeletal protein binding / Gene and protein expression by JAK-STAT signaling after Interleukin-12 stimulation / activation of protein kinase B activity / phosphotyrosine residue binding / cytoskeletal protein binding / Gene and protein expression by JAK-STAT signaling after Interleukin-12 stimulation / activation of protein kinase B activity / phosphotyrosine residue binding /  SH2 domain binding / SH2 domain binding /  ephrin receptor binding / cell chemotaxis / Downstream signal transduction / ephrin receptor binding / cell chemotaxis / Downstream signal transduction /  neutrophil chemotaxis / neutrophil chemotaxis /  protein tyrosine kinase binding / cellular response to nerve growth factor stimulus / negative regulation of protein phosphorylation / protein tyrosine kinase binding / cellular response to nerve growth factor stimulus / negative regulation of protein phosphorylation /  peptidylprolyl isomerase / peptidylprolyl isomerase /  peptidyl-prolyl cis-trans isomerase activity / positive regulation of protein secretion / Regulation of signaling by CBL / hippocampus development / regulation of actin cytoskeleton organization / FCGR3A-mediated phagocytosis / positive regulation of JNK cascade / negative regulation of protein kinase activity / Assembly Of The HIV Virion / peptidyl-prolyl cis-trans isomerase activity / positive regulation of protein secretion / Regulation of signaling by CBL / hippocampus development / regulation of actin cytoskeleton organization / FCGR3A-mediated phagocytosis / positive regulation of JNK cascade / negative regulation of protein kinase activity / Assembly Of The HIV Virion /  neuron migration / neuron migration /  neuromuscular junction / Budding and maturation of HIV virion / neuron differentiation / response to hydrogen peroxide / lipid metabolic process / neuromuscular junction / Budding and maturation of HIV virion / neuron differentiation / response to hydrogen peroxide / lipid metabolic process /  receptor tyrosine kinase binding / Regulation of actin dynamics for phagocytic cup formation / cerebral cortex development / receptor tyrosine kinase binding / Regulation of actin dynamics for phagocytic cup formation / cerebral cortex development /  platelet activation / platelet activation /  platelet aggregation / platelet aggregation /  kinase binding / VEGFA-VEGFR2 Pathway / kinase binding / VEGFA-VEGFR2 Pathway /  SH3 domain binding / : SH3 domain binding / : Similarity search - Function | ||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  SOLUTION NMR SOLUTION NMR | ||||||
| Model details | lowest energy, model1 | ||||||
 Authors Authors | Jankowski, W. / Saleh, T. / Rossi, P. / Kalodimos, C. | ||||||
 Citation Citation |  Journal: Nat.Chem.Biol. / Year: 2016 Journal: Nat.Chem.Biol. / Year: 2016Title: Cyclophilin A promotes cell migration via the Abl-Crk signaling pathway. Authors: Saleh, T. / Jankowski, W. / Sriram, G. / Rossi, P. / Shah, S. / Lee, K.B. / Cruz, L.A. / Rodriguez, A.J. / Birge, R.B. / Kalodimos, C.G. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2ms4.cif.gz 2ms4.cif.gz | 1.1 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2ms4.ent.gz pdb2ms4.ent.gz | 1000.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2ms4.json.gz 2ms4.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ms/2ms4 https://data.pdbj.org/pub/pdb/validation_reports/ms/2ms4 ftp://data.pdbj.org/pub/pdb/validation_reports/ms/2ms4 ftp://data.pdbj.org/pub/pdb/validation_reports/ms/2ms4 | HTTPS FTP |
|---|
-Related structure data
| Similar structure data | |
|---|---|
| Other databases |
- Links
Links
- Assembly
Assembly
| Deposited unit | 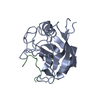
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 18036.504 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: PPIA, CYPA / Production host: Homo sapiens (human) / Gene: PPIA, CYPA / Production host:   Escherichia coli (E. coli) / References: UniProt: P62937, Escherichia coli (E. coli) / References: UniProt: P62937,  peptidylprolyl isomerase peptidylprolyl isomerase |
|---|---|
| #2: Protein/peptide |  Mass: 955.020 Da / Num. of mol.: 1 / Source method: obtained synthetically / References: UniProt: P46108*PLUS |
-Experimental details
-Experiment
| Experiment | Method:  SOLUTION NMR SOLUTION NMR | ||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details | Contents: 0.8 mM [U-100% 13C; U-100% 15N] protein_1, 90% H2O/10% D2O Solvent system: 90% H2O/10% D2O |
|---|---|
| Sample | Conc.: 0.8 mM / Component: entity_1-1 / Isotopic labeling: [U-100% 13C; U-100% 15N] |
| Sample conditions | Ionic strength: 0.1 / pH: 6.8 / Pressure: ambient / Temperature: 305 K |
-NMR measurement
| NMR spectrometer | Type: Bruker Avance / Manufacturer: Bruker / Model : AVANCE / Field strength: 700 MHz : AVANCE / Field strength: 700 MHz |
|---|
- Processing
Processing
| NMR software |
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| NMR representative | Selection criteria: lowest energy | |||||||||
| NMR ensemble | Conformer selection criteria: structures with the lowest energy Conformers calculated total number: 100 / Conformers submitted total number: 20 |
 Movie
Movie Controller
Controller













 PDBj
PDBj





















