[English] 日本語
 Yorodumi
Yorodumi- PDB-2lnf: Neurotensin 40 structures in DMPC/CHAPS(q=0.25) bicelle pH 5.5 & ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2lnf | ||||||
|---|---|---|---|---|---|---|---|
| Title | Neurotensin 40 structures in DMPC/CHAPS(q=0.25) bicelle pH 5.5 & 298K. NMR data & Structures | ||||||
 Components Components | Neurotensin | ||||||
 Keywords Keywords |  NEUROPEPTIDE / NT / DMPC:CHAPS Bicelle NEUROPEPTIDE / NT / DMPC:CHAPS Bicelle | ||||||
| Function / homology |  Function and homology information Function and homology information neuropeptide receptor binding / neuropeptide hormone activity / neuropeptide signaling pathway / axon terminus / neuropeptide receptor binding / neuropeptide hormone activity / neuropeptide signaling pathway / axon terminus /  transport vesicle / blood vessel diameter maintenance / Peptide ligand-binding receptors / positive regulation of NF-kappaB transcription factor activity / G alpha (q) signalling events / transport vesicle / blood vessel diameter maintenance / Peptide ligand-binding receptors / positive regulation of NF-kappaB transcription factor activity / G alpha (q) signalling events /  receptor ligand activity ... receptor ligand activity ... neuropeptide receptor binding / neuropeptide hormone activity / neuropeptide signaling pathway / axon terminus / neuropeptide receptor binding / neuropeptide hormone activity / neuropeptide signaling pathway / axon terminus /  transport vesicle / blood vessel diameter maintenance / Peptide ligand-binding receptors / positive regulation of NF-kappaB transcription factor activity / G alpha (q) signalling events / transport vesicle / blood vessel diameter maintenance / Peptide ligand-binding receptors / positive regulation of NF-kappaB transcription factor activity / G alpha (q) signalling events /  receptor ligand activity / positive regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction / negative regulation of gene expression / intracellular membrane-bounded organelle / positive regulation of gene expression / receptor ligand activity / positive regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction / negative regulation of gene expression / intracellular membrane-bounded organelle / positive regulation of gene expression /  signal transduction / extracellular region signal transduction / extracellular regionSimilarity search - Function | ||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  SOLUTION NMR / SOLUTION NMR /  simulated annealing simulated annealing | ||||||
| Model details | lowest energy, model 1 | ||||||
 Authors Authors | Mukhopadhyay, C. / Khatun, U. | ||||||
 Citation Citation |  Journal: Biophys.Chem. / Year: 2012 Journal: Biophys.Chem. / Year: 2012Title: Modulation of the neurotensin solution structure in the presence of ganglioside GM1 bicelle. Authors: Khatun, U.L. / Goswami, S.K. / Mukhopadhyay, C. | ||||||
| History |
|
- Structure visualization
Structure visualization
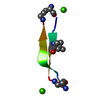
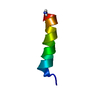
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2lnf.cif.gz 2lnf.cif.gz | 167.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2lnf.ent.gz pdb2lnf.ent.gz | 119.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2lnf.json.gz 2lnf.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ln/2lnf https://data.pdbj.org/pub/pdb/validation_reports/ln/2lnf ftp://data.pdbj.org/pub/pdb/validation_reports/ln/2lnf ftp://data.pdbj.org/pub/pdb/validation_reports/ln/2lnf | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein/peptide |  / NT / NTMass: 1693.964 Da / Num. of mol.: 1 / Fragment: UNP RESIDUES 151-163 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: NTS / Production host: Homo sapiens (human) / Gene: NTS / Production host:   Escherichia coli (E. coli) / References: UniProt: P30990 Escherichia coli (E. coli) / References: UniProt: P30990 |
|---|
-Experimental details
-Experiment
| Experiment | Method:  SOLUTION NMR SOLUTION NMR |
|---|---|
| NMR experiment | Type : 2D 1H-1H TOCSY, 2D 1H-1H ROESY,2D DQF- : 2D 1H-1H TOCSY, 2D 1H-1H ROESY,2D DQF- COSY COSY |
- Sample preparation
Sample preparation
| Details | Contents: 4.5 mM Neurotensin-1, 90% H2O/10% D2O / Solvent system: 90% H2O/10% D2O |
|---|---|
| Sample | Conc.: 4.5 mM / Component: Neurotensin-1 |
| Sample conditions | Ionic strength: 4.5 / pH: 5.5 / Pressure: ambient / Temperature: 298 K |
-NMR measurement
| NMR spectrometer | Type: Bruker DRX / Manufacturer: Bruker / Model : DRX / Field strength: 500 MHz : DRX / Field strength: 500 MHz |
|---|
- Processing
Processing
| NMR software |
| |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method:  simulated annealing / Software ordinal: 1 simulated annealing / Software ordinal: 1 | |||||||||||||||||||||
| NMR representative | Selection criteria: lowest energy | |||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the lowest energy Conformers calculated total number: 100 / Conformers submitted total number: 40 / Representative conformer: 1 |
 Movie
Movie Controller
Controller










 PDBj
PDBj




