+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2l41 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Nab3 RRM - UCUU complex | ||||||
 Components Components |
| ||||||
 Keywords Keywords | RNA BINDING PROTEIN/RNA / Nab3 RRM / UCUU oligonucleotide / RNA BINDING PROTEIN-RNA complex | ||||||
| Function / homology |  Function and homology information Function and homology informationtranscription regulatory region RNA binding / antisense RNA transcript catabolic process /  Nrd1 complex / sno(s)RNA 3'-end processing / termination of RNA polymerase II transcription, exosome-dependent / snRNA 3'-end processing / tRNA 3'-end processing / CUT catabolic process / : / nuclear mRNA surveillance ...transcription regulatory region RNA binding / antisense RNA transcript catabolic process / Nrd1 complex / sno(s)RNA 3'-end processing / termination of RNA polymerase II transcription, exosome-dependent / snRNA 3'-end processing / tRNA 3'-end processing / CUT catabolic process / : / nuclear mRNA surveillance ...transcription regulatory region RNA binding / antisense RNA transcript catabolic process /  Nrd1 complex / sno(s)RNA 3'-end processing / termination of RNA polymerase II transcription, exosome-dependent / snRNA 3'-end processing / tRNA 3'-end processing / CUT catabolic process / : / nuclear mRNA surveillance / : / mRNA 3'-end processing / Processing of Capped Intron-Containing Pre-mRNA / Nrd1 complex / sno(s)RNA 3'-end processing / termination of RNA polymerase II transcription, exosome-dependent / snRNA 3'-end processing / tRNA 3'-end processing / CUT catabolic process / : / nuclear mRNA surveillance / : / mRNA 3'-end processing / Processing of Capped Intron-Containing Pre-mRNA /  mRNA binding / mRNA binding /  RNA binding / RNA binding /  nucleoplasm / nucleoplasm /  nucleus / nucleus /  cytoplasm cytoplasmSimilarity search - Function | ||||||
| Biological species |   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast) | ||||||
| Method |  SOLUTION NMR / SOLUTION NMR /  molecular dynamics molecular dynamics | ||||||
| Model details | lowest energy, model 1 | ||||||
 Authors Authors | Stefl, R. / Pergoli, R. / Hobor, F. / Kubicek, K. / Zimmermann, M. / Pasulka, J. / Hofr, C. | ||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 2011 Journal: J.Biol.Chem. / Year: 2011Title: Recognition of transcription termination signal by the nuclear polyadenylated RNA-binding (NAB) 3 protein Authors: Hobor, F. / Pergoli, R. / Kubicek, K. / Hrossova, D. / Bacikova, V. / Zimmermann, M. / Pasulka, J. / Hofr, C. / Vanacova, S. / Stefl, R. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2l41.cif.gz 2l41.cif.gz | 528.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2l41.ent.gz pdb2l41.ent.gz | 447.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2l41.json.gz 2l41.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/l4/2l41 https://data.pdbj.org/pub/pdb/validation_reports/l4/2l41 ftp://data.pdbj.org/pub/pdb/validation_reports/l4/2l41 ftp://data.pdbj.org/pub/pdb/validation_reports/l4/2l41 | HTTPS FTP |
|---|
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 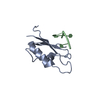
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein |  RNA recognition motif / RRM domain from Nab3 RNA recognition motif / RRM domain from Nab3Mass: 8829.131 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast)Gene: NAB3 / Production host:   Escherichia coli (E. coli) / References: UniProt: P38996 Escherichia coli (E. coli) / References: UniProt: P38996 |
|---|---|
| #2: RNA chain | Mass: 1178.722 Da / Num. of mol.: 1 / Source method: obtained synthetically |
-Experimental details
-Experiment
| Experiment | Method:  SOLUTION NMR SOLUTION NMR | ||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details | Contents: 2.5mM [U-99% 13C; U-99% 15N] Nab3 RRM-1, 2.5mM RNA-2, 50mM sodium phosphate buffer(pH 7.5)-3, 150mM sodium chloride-4, 10mM beta-mercaptoethanol-5, 90% H2O/10% D2O Solvent system: 90% H2O/10% D2O | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample |
| ||||||||||||||||||||||||
| Sample conditions | pH: 7.5 / Pressure: ambient / Temperature: 303 K |
-NMR measurement
| NMR spectrometer |
|
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method:  molecular dynamics / Software ordinal: 1 / Details: Structures refined in explicit solvent molecular dynamics / Software ordinal: 1 / Details: Structures refined in explicit solvent | ||||||||||||||||||||||||||||||||||||||||||||
| NMR representative | Selection criteria: lowest energy | ||||||||||||||||||||||||||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the lowest energy Conformers calculated total number: 60 / Conformers submitted total number: 20 / Representative conformer: 1 |
 Movie
Movie Controller
Controller













 PDBj
PDBj
