[English] 日本語
 Yorodumi
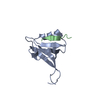
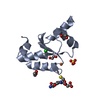
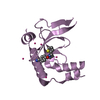
Yorodumi- PDB-2l1a: Solution NMR structure of the N-terminal GTPase-like domain of di... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2l1a | ||||||
|---|---|---|---|---|---|---|---|
| Title | Solution NMR structure of the N-terminal GTPase-like domain of dictyostelium discoideum Fomin C | ||||||
 Components Components | Formin-C | ||||||
 Keywords Keywords | actin binding protein / fruiting body formation | ||||||
| Function / homology |  Function and homology information Function and homology informationsorocarp spore cell differentiation /  macropinosome / actin filament network formation / culmination involved in sorocarp development / cortical actin cytoskeleton organization / actin filament bundle assembly / macropinosome / actin filament network formation / culmination involved in sorocarp development / cortical actin cytoskeleton organization / actin filament bundle assembly /  actin filament binding / cell-cell junction / actin filament binding / cell-cell junction /  gene expression / gene expression /  cytoskeleton ...sorocarp spore cell differentiation / cytoskeleton ...sorocarp spore cell differentiation /  macropinosome / actin filament network formation / culmination involved in sorocarp development / cortical actin cytoskeleton organization / actin filament bundle assembly / macropinosome / actin filament network formation / culmination involved in sorocarp development / cortical actin cytoskeleton organization / actin filament bundle assembly /  actin filament binding / cell-cell junction / actin filament binding / cell-cell junction /  gene expression / gene expression /  cytoskeleton / cytoskeleton /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function | ||||||
| Biological species |   Dictyostelium discoideum (eukaryote) Dictyostelium discoideum (eukaryote) | ||||||
| Method |  SOLUTION NMR / torsion angle dynamics, SOLUTION NMR / torsion angle dynamics,  simulated annealing, final refinement in water-shell simulated annealing, final refinement in water-shell | ||||||
| Model details | lowest energy, model 1 | ||||||
 Authors Authors | Dames, S.A. / Schoenichen, A. / Stephan, G. / Geyer, M. | ||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 2011 Journal: J.Biol.Chem. / Year: 2011Title: Structure, dynamics, lipid binding, and physiological relevance of the putative GTPase-binding domain of Dictyostelium formin C. Authors: Dames, S.A. / Junemann, A. / Sass, H.J. / Schonichen, A. / Stopschinski, B.E. / Grzesiek, S. / Faix, J. / Geyer, M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2l1a.cif.gz 2l1a.cif.gz | 661.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2l1a.ent.gz pdb2l1a.ent.gz | 551.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2l1a.json.gz 2l1a.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/l1/2l1a https://data.pdbj.org/pub/pdb/validation_reports/l1/2l1a ftp://data.pdbj.org/pub/pdb/validation_reports/l1/2l1a ftp://data.pdbj.org/pub/pdb/validation_reports/l1/2l1a | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 12075.733 Da / Num. of mol.: 1 Fragment: N-terminal potential GTPase-binding domain, residues 1-100 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Dictyostelium discoideum (eukaryote) / Gene: forC, DDB_G0287295 / Plasmid: pGEX-4T1 tev / Production host: Dictyostelium discoideum (eukaryote) / Gene: forC, DDB_G0287295 / Plasmid: pGEX-4T1 tev / Production host:   Escherichia coli (E. coli) / References: UniProt: Q54KF1 Escherichia coli (E. coli) / References: UniProt: Q54KF1 |
|---|
-Experimental details
-Experiment
| Experiment | Method:  SOLUTION NMR SOLUTION NMR | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sample conditions | Ionic strength: 0.1 / pH: 6.5 / Pressure: ambient / Temperature: 293 K |
-NMR measurement
| NMR spectrometer |
|
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: torsion angle dynamics,  simulated annealing, final refinement in water-shell simulated annealing, final refinement in water-shellSoftware ordinal: 1 | ||||||||||||||||||
| NMR representative | Selection criteria: lowest energy | ||||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the lowest energy Conformers calculated total number: 200 / Conformers submitted total number: 20 |
 Movie
Movie Controller
Controller









 PDBj
PDBj