[English] 日本語
 Yorodumi
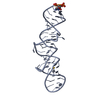
Yorodumi- PDB-2klr: Solid-state NMR structure of the alpha-crystallin domain in alpha... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2klr | ||||||
|---|---|---|---|---|---|---|---|
| Title | Solid-state NMR structure of the alpha-crystallin domain in alphaB-crystallin oligomers | ||||||
 Components Components | Alpha-crystallin B chain CRYAB CRYAB | ||||||
 Keywords Keywords |  CHAPERONE / CHAPERONE /  Protein / dimer / Protein / dimer /  oligomer / oligomer /  heterogeneity / intermolecular interactions / sHSP / heterogeneity / intermolecular interactions / sHSP /  human / small heat-shock protein / human / small heat-shock protein /  Cataract / Desmin-related myopathy / Disease mutation / Cataract / Desmin-related myopathy / Disease mutation /  Eye lens protein / Eye lens protein /  Glycoprotein / Glycoprotein /  Methylation / Methylation /  Oxidation / Oxidation /  Phosphoprotein Phosphoprotein | ||||||
| Function / homology |  Function and homology information Function and homology informationmicrotubule polymerization or depolymerization / negative regulation of intracellular transport / apoptotic process involved in morphogenesis / cardiac myofibril /  regulation of programmed cell death / tubulin complex assembly / structural constituent of eye lens / negative regulation of amyloid fibril formation / regulation of programmed cell death / tubulin complex assembly / structural constituent of eye lens / negative regulation of amyloid fibril formation /  M band / lens development in camera-type eye ...microtubule polymerization or depolymerization / negative regulation of intracellular transport / apoptotic process involved in morphogenesis / cardiac myofibril / M band / lens development in camera-type eye ...microtubule polymerization or depolymerization / negative regulation of intracellular transport / apoptotic process involved in morphogenesis / cardiac myofibril /  regulation of programmed cell death / tubulin complex assembly / structural constituent of eye lens / negative regulation of amyloid fibril formation / regulation of programmed cell death / tubulin complex assembly / structural constituent of eye lens / negative regulation of amyloid fibril formation /  M band / lens development in camera-type eye / muscle organ development / actin filament bundle / HSF1-dependent transactivation / negative regulation of reactive oxygen species metabolic process / negative regulation of protein-containing complex assembly / stress-activated MAPK cascade / M band / lens development in camera-type eye / muscle organ development / actin filament bundle / HSF1-dependent transactivation / negative regulation of reactive oxygen species metabolic process / negative regulation of protein-containing complex assembly / stress-activated MAPK cascade /  synaptic membrane / synaptic membrane /  muscle contraction / cellular response to gamma radiation / response to hydrogen peroxide / negative regulation of cell growth / Z disc / unfolded protein binding / muscle contraction / cellular response to gamma radiation / response to hydrogen peroxide / negative regulation of cell growth / Z disc / unfolded protein binding /  protein folding / response to estradiol / protein folding / response to estradiol /  amyloid-beta binding / response to heat / amyloid-beta binding / response to heat /  perikaryon / protein refolding / perikaryon / protein refolding /  microtubule binding / microtubule binding /  dendritic spine / dendritic spine /  lysosome / response to hypoxia / protein stabilization / lysosome / response to hypoxia / protein stabilization /  axon / negative regulation of gene expression / negative regulation of DNA-templated transcription / protein-containing complex binding / negative regulation of apoptotic process / structural molecule activity / axon / negative regulation of gene expression / negative regulation of DNA-templated transcription / protein-containing complex binding / negative regulation of apoptotic process / structural molecule activity /  cell surface / protein homodimerization activity / protein-containing complex / cell surface / protein homodimerization activity / protein-containing complex /  mitochondrion / extracellular exosome / mitochondrion / extracellular exosome /  nucleoplasm / identical protein binding / nucleoplasm / identical protein binding /  metal ion binding / metal ion binding /  nucleus / nucleus /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function | ||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||
| Method | SOLID-STATE NMR /  simulated annealing, simulated annealing,  molecular dynamics, torsion angle dynamics molecular dynamics, torsion angle dynamics | ||||||
| Model details | lowest energy, model 1 | ||||||
 Authors Authors | Jehle, S. / Rajagopal, P. / Markovic, S. / Bardiaux, B. / Kuehne, R. / Higman, V.A. / Klevit, R.E. / van Rossum, B. / Oschkinat, H. | ||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2010 Journal: Nat Struct Mol Biol / Year: 2010Title: Solid-state NMR and SAXS studies provide a structural basis for the activation of alphaB-crystallin oligomers. Authors: Stefan Jehle / Ponni Rajagopal / Benjamin Bardiaux / Stefan Markovic / Ronald Kühne / Joseph R Stout / Victoria A Higman / Rachel E Klevit / Barth-Jan van Rossum / Hartmut Oschkinat /  Abstract: The small heat shock protein alphaB-crystallin (alphaB) contributes to cellular protection against stress. For decades, high-resolution structural studies on oligomeric alphaB have been confounded by ...The small heat shock protein alphaB-crystallin (alphaB) contributes to cellular protection against stress. For decades, high-resolution structural studies on oligomeric alphaB have been confounded by its polydisperse nature. Here, we present a structural basis of oligomer assembly and activation of the chaperone using solid-state NMR and small-angle X-ray scattering (SAXS). The basic building block is a curved dimer, with an angle of approximately 121 degrees between the planes of the beta-sandwich formed by alpha-crystallin domains. The highly conserved IXI motif covers a substrate binding site at pH 7.5. We observe a pH-dependent modulation of the interaction of the IXI motif with beta4 and beta8, consistent with a pH-dependent regulation of the chaperone function. N-terminal region residues Ser59-Trp60-Phe61 are involved in intermolecular interaction with beta3. Intermolecular restraints from NMR and volumetric restraints from SAXS were combined to calculate a model of a 24-subunit alphaB oligomer with tetrahedral symmetry. #1: Journal: J.Mol.Biol. / Year: 2009 Title: AlphaB-crystallin: a hybrid solid-state/solution-state NMR investigation reveals structural aspects of the heterogeneous oligomer. Authors: Jehle, S. / van Rossum, B. / Stout, J.R. / Noguchi, S.M. / Falber, K. / Rehbein, K. / Oschkinat, H. / Klevit, R.E. / Rajagopal, P. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2klr.cif.gz 2klr.cif.gz | 528.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2klr.ent.gz pdb2klr.ent.gz | 448.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2klr.json.gz 2klr.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/kl/2klr https://data.pdbj.org/pub/pdb/validation_reports/kl/2klr ftp://data.pdbj.org/pub/pdb/validation_reports/kl/2klr ftp://data.pdbj.org/pub/pdb/validation_reports/kl/2klr | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||||||||
| NMR ensembles |
| ||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS oper:
| ||||||||||||||||
| Details | The authors state that the NCS Matrix for the hexamer should be applied to model 7 in the ensemble. |
- Components
Components
| #1: Protein |  CRYAB / Alpha(B)-crystallin / Rosenthal fiber component / Heat shock protein beta-5 / HspB5 / Renal ...Alpha(B)-crystallin / Rosenthal fiber component / Heat shock protein beta-5 / HspB5 / Renal carcinoma antigen NY-REN-27 CRYAB / Alpha(B)-crystallin / Rosenthal fiber component / Heat shock protein beta-5 / HspB5 / Renal ...Alpha(B)-crystallin / Rosenthal fiber component / Heat shock protein beta-5 / HspB5 / Renal carcinoma antigen NY-REN-27Mass: 20191.930 Da / Num. of mol.: 2 Fragment: Alpha-crystallin domain homodimer in oligomers of alphaB-crystallin Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: CRYAB, CRYA2 / Production host: Homo sapiens (human) / Gene: CRYAB, CRYA2 / Production host:   Escherichia coli (E. coli) / References: UniProt: P02511 Escherichia coli (E. coli) / References: UniProt: P02511Sequence details | THE HOMOGENEOUS PART OF THE ALPHA-CRYSTALLIN DOMAIN DIMER IN OLIGOMERS OF HUMAN ALPHAB CRYSTALLIN ...THE HOMOGENEOU | |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLID-STATE NMR | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details |
| ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample |
| ||||||||||||||||||||||||||||||||||||
| Sample conditions | pH: 7.6 / Temperature: 270 K |
-NMR measurement
| NMR spectrometer |
|
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method:  simulated annealing, simulated annealing,  molecular dynamics, torsion angle dynamics molecular dynamics, torsion angle dynamicsSoftware ordinal: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||
| NMR representative | Selection criteria: lowest energy | ||||||||||||||||||||||||||||||||||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the lowest energy Conformers calculated total number: 200 / Conformers submitted total number: 10 |
 Movie
Movie Controller
Controller











 PDBj
PDBj


