[English] 日本語
 Yorodumi
Yorodumi- PDB-2fz5: Solution structure of two-electron reduced Megasphaera elsdenii f... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2fz5 | ||||||
|---|---|---|---|---|---|---|---|
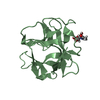
| Title | Solution structure of two-electron reduced Megasphaera elsdenii flavodoxin | ||||||
 Components Components | Flavodoxin | ||||||
 Keywords Keywords |  ELECTRON TRANSPORT / alpha/beta doubly-wound topology / non-covalently bound FMN ELECTRON TRANSPORT / alpha/beta doubly-wound topology / non-covalently bound FMN | ||||||
| Function / homology |  Function and homology information Function and homology information | ||||||
| Biological species |  Megasphaera elsdenii (bacteria) Megasphaera elsdenii (bacteria) | ||||||
| Method |  SOLUTION NMR / restrained molecular dynamics SOLUTION NMR / restrained molecular dynamics | ||||||
 Authors Authors | van Mierlo, C.P.M. / Lijnzaad, P. / Vervoort, J. / Mueller, F. / Berendsen, H.J. / de Vlieg, J. | ||||||
 Citation Citation |  Journal: Eur.J.Biochem. / Year: 1990 Journal: Eur.J.Biochem. / Year: 1990Title: Tertiary structure of two-electron reduced Megasphaera elsdenii flavodoxin and some implications, as determined by two-dimensional 1H NMR and restrained molecular dynamics Authors: van Mierlo, C.P.M. / Lijnzaad, P. / Vervoort, J. / Mueller, F. / Berendsen, H.J. / de Vlieg, J. #1: Journal: Eur.J.Biochem. / Year: 1990 Title: Secondary and tertiary structure characteristics of Megasphaera elsdenii flavodoxin in the reduced state as determined by two-dimensional 1H NMR Authors: van Mierlo, C.P.M. / Mueller, F. / Vervoort, J. #2: Journal: Eur.J.Biochem. / Year: 1990 Title: A two-dimensional 1H NMR study on Megasphaera elsdenii flavodoxin in the reduced state: sequential assignments Authors: van Mierlo, C.P.M. / Vervoort, J. / Mueller, F. / Bacher, A. #3: Journal: Eur.J.Biochem. / Year: 1990 Title: A two-dimensional 1H NMR study on Megasphaera elsdenii flavodoxin in the oxidized state and some comparisons with the two-electron reduced state Authors: van Mierlo, C.P.M. / van der Sanden, B.P. / van Woensel, P. / Mueller, F. / Vervoort, J. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2fz5.cif.gz 2fz5.cif.gz | 38.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2fz5.ent.gz pdb2fz5.ent.gz | 26.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2fz5.json.gz 2fz5.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/fz/2fz5 https://data.pdbj.org/pub/pdb/validation_reports/fz/2fz5 ftp://data.pdbj.org/pub/pdb/validation_reports/fz/2fz5 ftp://data.pdbj.org/pub/pdb/validation_reports/fz/2fz5 | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein |  Mass: 14561.183 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Megasphaera elsdenii (bacteria) / References: UniProt: P00321 Megasphaera elsdenii (bacteria) / References: UniProt: P00321 |
|---|---|
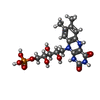
| #2: Chemical | ChemComp-FNR / |
-Experimental details
-Experiment
| Experiment | Method:  SOLUTION NMR SOLUTION NMR | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||||||||||||||
| NMR details | Text: NOESY spectra had mixing times 50, 100, 150 and 200 ms. Double Quantum spectra had a delay of 32 ms. Homonuclear Hartmann Hahn transfer spectra (using MLEV-17 composite pulse cycling) had mixing times 10-160 ms |
- Sample preparation
Sample preparation
| Details |
| ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample conditions |
|
-NMR measurement
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Radiation wavelength | Relative weight: 1 | ||||||||||||||||||||
| NMR spectrometer |
|
- Processing
Processing
| NMR software |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: restrained molecular dynamics / Software ordinal: 1 Details: The Xray structure of the semiquinone state of Clostridium MP flavodoxin was used as a starting structure for restrained molecular dynamics (RMD) calculations in vacuo (van Mierlo et al. Eur. ...Details: The Xray structure of the semiquinone state of Clostridium MP flavodoxin was used as a starting structure for restrained molecular dynamics (RMD) calculations in vacuo (van Mierlo et al. Eur. J. Biochem. 194, 185 - 198 (1990)). 509 distance restraints, 293 medium, 216 long-range were used in refinement. One repulsive restraint, between N5H of FMN and NH of E60 was included. RMD run was of 120 ps. During the first 50 ps the force constant was gradually increased to a high value of 4000 kJmol-1nm-2. From 50 ps on, the force constant was kept constant. The time span of 60-120 ps, at a time resolution of 0.02 ps, was used for calculating the average structure. The time-averaged (not energy minimised) structure has a potential energy -2278 +/- 122 kJmol-1, time-averaged sum of all violations is 2.27 nm, largest occuring violation is 66 pm | ||||||||||||
| NMR representative | Selection criteria: averaging the restrained molecular dynamics trajectory in the range 60-120 ps. | ||||||||||||
| NMR ensemble | Conformers calculated total number: 3000 / Conformers submitted total number: 1 |
 Movie
Movie Controller
Controller










 PDBj
PDBj