[English] 日本語
 Yorodumi
Yorodumi- PDB-2fo5: Crystal structure of recombinant barley cysteine endoprotease B i... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2fo5 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of recombinant barley cysteine endoprotease B isoform 2 (EP-B2) in complex with leupeptin | ||||||
 Components Components |
| ||||||
 Keywords Keywords | HYDROLASE/HYDROLASE INHIBITOR / EP-B2 / EPB2 / EPB / cysteine endoprotease /  endopeptidase / endopeptidase /  leupeptin / leupeptin /  HYDROLASE / HYDROLASE-HYDROLASE INHIBITOR COMPLEX HYDROLASE / HYDROLASE-HYDROLASE INHIBITOR COMPLEX | ||||||
| Function / homology |  Function and homology information Function and homology informationcysteine-type peptidase activity /  Hydrolases; Acting on peptide bonds (peptidases); Cysteine endopeptidases Hydrolases; Acting on peptide bonds (peptidases); Cysteine endopeptidasesSimilarity search - Function | ||||||
| Biological species |   Hordeum vulgare (barley) Hordeum vulgare (barley) Actinomycetes Streptomyces roseus MA 839-A1 (bacteria) Actinomycetes Streptomyces roseus MA 839-A1 (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.2 Å MOLECULAR REPLACEMENT / Resolution: 2.2 Å | ||||||
 Authors Authors | Bethune, M.T. / Strop, P. / Brunger, A.T. / Khosla, C. | ||||||
 Citation Citation |  Journal: Chem.Biol. / Year: 2006 Journal: Chem.Biol. / Year: 2006Title: Heterologous Expression, Purification, Refolding, and Structural-Functional Characterization of EP-B2, a Self-Activating Barley Cysteine Endoprotease. Authors: Bethune, M.T. / Strop, P. / Tang, Y. / Sollid, L.M. / Khosla, C. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2fo5.cif.gz 2fo5.cif.gz | 188 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2fo5.ent.gz pdb2fo5.ent.gz | 155.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2fo5.json.gz 2fo5.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/fo/2fo5 https://data.pdbj.org/pub/pdb/validation_reports/fo/2fo5 ftp://data.pdbj.org/pub/pdb/validation_reports/fo/2fo5 ftp://data.pdbj.org/pub/pdb/validation_reports/fo/2fo5 | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| 3 | 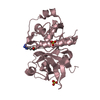
| ||||||||
| 4 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein |  Mass: 27759.693 Da / Num. of mol.: 4 / Fragment: Cystein proteinase EP-B2 domain, residues 133-356 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Hordeum vulgare (barley) / Gene: EPB2 / Plasmid: pMTB1 / Species (production host): Escherichia coli / Production host: Hordeum vulgare (barley) / Gene: EPB2 / Plasmid: pMTB1 / Species (production host): Escherichia coli / Production host:   Escherichia coli BL21 (bacteria) / Strain (production host): BL21 Escherichia coli BL21 (bacteria) / Strain (production host): BL21References: UniProt: P25250,  Hydrolases; Acting on peptide bonds (peptidases); Cysteine endopeptidases Hydrolases; Acting on peptide bonds (peptidases); Cysteine endopeptidases#2: Protein/peptide |  #3: Chemical | ChemComp-SO4 /  Sulfate Sulfate#4: Water | ChemComp-HOH / |  Water WaterCompound details | THE LEUPEPTIN IS COVALENTLY | Sequence details | THERE IS A DISCREPANCY IN THE NORINE AND PDB NUMBERING, AS NORINE COUNTS ACE AND LEU TOGETHER AS ...THERE IS A DISCREPANC | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.2 Å3/Da / Density % sol: 44 % |
|---|---|
Crystal grow | Temperature: 298 K / Method: vapor diffusion, hanging drop / pH: 7 Details: 1.4 M LiSO4 0.1 M HEPES, pH 7.0, VAPOR DIFFUSION, HANGING DROP, temperature 298K |
-Data collection
| Diffraction | Mean temperature: 113 K | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ALS ALS  / Beamline: 8.3.1 / Wavelength: 1.115869 Å / Beamline: 8.3.1 / Wavelength: 1.115869 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Detector | Type: ADSC QUANTUM 4 / Detector: CCD / Date: Oct 22, 2005 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength : 1.115869 Å / Relative weight: 1 : 1.115869 Å / Relative weight: 1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection | Resolution: 2.15→50 Å / Num. obs: 48898 / % possible obs: 89.9 % / Redundancy: 3.3 % / Rmerge(I) obs: 0.059 / Χ2: 1.181 / Net I/σ(I): 11.7 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection shell |
|
-Phasing
| Phasing MR |
|
|---|
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT / Resolution: 2.2→15 Å / FOM work R set: 0.843 / σ(F): 0 / Stereochemistry target values: Engh & Huber MOLECULAR REPLACEMENT / Resolution: 2.2→15 Å / FOM work R set: 0.843 / σ(F): 0 / Stereochemistry target values: Engh & Huber
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Bsol: 44.175 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 26.942 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.2→15 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Total num. of bins used: 50
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Xplor file |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj