[English] 日本語
 Yorodumi
Yorodumi- PDB-1tf0: Crystal structure of the GA module complexed with human serum albumin -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1tf0 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of the GA module complexed with human serum albumin | ||||||
 Components Components |
| ||||||
 Keywords Keywords |  PROTEIN BINDING / PROTEIN-PROTEIN COMPLEX PROTEIN BINDING / PROTEIN-PROTEIN COMPLEX | ||||||
| Function / homology |  Function and homology information Function and homology informationcellular response to calcium ion starvation /  exogenous protein binding / Ciprofloxacin ADME / exogenous protein binding / Ciprofloxacin ADME /  enterobactin binding / enterobactin binding /  Heme biosynthesis / HDL remodeling / negative regulation of mitochondrial depolarization / Prednisone ADME / Heme degradation / Aspirin ADME ...cellular response to calcium ion starvation / Heme biosynthesis / HDL remodeling / negative regulation of mitochondrial depolarization / Prednisone ADME / Heme degradation / Aspirin ADME ...cellular response to calcium ion starvation /  exogenous protein binding / Ciprofloxacin ADME / exogenous protein binding / Ciprofloxacin ADME /  enterobactin binding / enterobactin binding /  Heme biosynthesis / HDL remodeling / negative regulation of mitochondrial depolarization / Prednisone ADME / Heme degradation / Aspirin ADME / Heme biosynthesis / HDL remodeling / negative regulation of mitochondrial depolarization / Prednisone ADME / Heme degradation / Aspirin ADME /  antioxidant activity / antioxidant activity /  toxic substance binding / toxic substance binding /  small molecule binding / Scavenging of heme from plasma / Recycling of bile acids and salts / cellular response to starvation / platelet alpha granule lumen / small molecule binding / Scavenging of heme from plasma / Recycling of bile acids and salts / cellular response to starvation / platelet alpha granule lumen /  fatty acid binding / fatty acid binding /  Post-translational protein phosphorylation / Cytoprotection by HMOX1 / Regulation of Insulin-like Growth Factor (IGF) transport and uptake by Insulin-like Growth Factor Binding Proteins (IGFBPs) / Post-translational protein phosphorylation / Cytoprotection by HMOX1 / Regulation of Insulin-like Growth Factor (IGF) transport and uptake by Insulin-like Growth Factor Binding Proteins (IGFBPs) /  pyridoxal phosphate binding / Platelet degranulation / protein-folding chaperone binding / blood microparticle / copper ion binding / pyridoxal phosphate binding / Platelet degranulation / protein-folding chaperone binding / blood microparticle / copper ion binding /  endoplasmic reticulum lumen / endoplasmic reticulum lumen /  Golgi apparatus / Golgi apparatus /  endoplasmic reticulum / protein-containing complex / endoplasmic reticulum / protein-containing complex /  DNA binding / DNA binding /  extracellular space / extracellular exosome / extracellular region / identical protein binding / extracellular space / extracellular exosome / extracellular region / identical protein binding /  nucleus / nucleus /  cytoplasm cytoplasmSimilarity search - Function | ||||||
| Biological species |   Finegoldia magna (bacteria) Finegoldia magna (bacteria)  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.7 Å MOLECULAR REPLACEMENT / Resolution: 2.7 Å | ||||||
 Authors Authors | Lejon, S. / Svensson, S. / Bjorck, L. / Frick, I.-M. / Wikstrom, M. | ||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 2004 Journal: J.Biol.Chem. / Year: 2004Title: Crystal structure and biological implications of a bacterial albumin binding module in complex with human serum albumin Authors: Lejon, S. / Frick, I.-M. / Bjorck, L. / Wikstrom, M. / Svensson, S. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1tf0.cif.gz 1tf0.cif.gz | 132.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1tf0.ent.gz pdb1tf0.ent.gz | 103.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1tf0.json.gz 1tf0.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/tf/1tf0 https://data.pdbj.org/pub/pdb/validation_reports/tf/1tf0 ftp://data.pdbj.org/pub/pdb/validation_reports/tf/1tf0 ftp://data.pdbj.org/pub/pdb/validation_reports/tf/1tf0 | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein |  Mass: 65317.680 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Homo sapiens (human) / Tissue fraction: plasma / References: UniProt: P02768 Homo sapiens (human) / Tissue fraction: plasma / References: UniProt: P02768 | ||
|---|---|---|---|
| #2: Protein | Mass: 5939.786 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Finegoldia magna (bacteria) / Strain: ATCC 29328 / Gene: PAB / Plasmid: pHD389 / Production host: Finegoldia magna (bacteria) / Strain: ATCC 29328 / Gene: PAB / Plasmid: pHD389 / Production host:   Escherichia coli (E. coli) / Strain (production host): JM109 / References: UniProt: Q51911 Escherichia coli (E. coli) / Strain (production host): JM109 / References: UniProt: Q51911 | ||
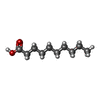
| #3: Chemical |  Capric acid Capric acid#4: Chemical | ChemComp-CIT / |  Citric acid Citric acid |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.7 Å3/Da / Density % sol: 55 % |
|---|---|
Crystal grow | Temperature: 291 K / Method: vapor diffusion, hanging drop / pH: 6 Details: Ammonium sulfate, citrate, pH 6.0, VAPOR DIFFUSION, HANGING DROP, temperature 291K |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: ID14-4 / Wavelength: 1 Å / Beamline: ID14-4 / Wavelength: 1 Å |
| Detector | Type: ADSC QUANTUM 4 / Detector: CCD / Date: Sep 15, 2003 / Details: Toroidal mirror |
| Radiation | Monochromator: Double crystal, Si(111) or Si(311) / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 1 Å / Relative weight: 1 : 1 Å / Relative weight: 1 |
| Reflection | Resolution: 2.6→100 Å / Num. all: 24858 / Num. obs: 24858 / Observed criterion σ(F): 5 / Observed criterion σ(I): 5 / Redundancy: 10 % / Rmerge(I) obs: 0.07 / Net I/σ(I): 13.1 |
| Reflection shell | Resolution: 2.6→2.64 Å / Rmerge(I) obs: 0.374 / Mean I/σ(I) obs: 4.4 / % possible all: 100 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB entries 1AO6 and 1PRB Resolution: 2.7→65 Å / Cor.coef. Fo:Fc: 0.916 / Cor.coef. Fo:Fc free: 0.879 / SU B: 31.497 / SU ML: 0.299 / TLS residual ADP flag: LIKELY RESIDUAL / Cross valid method: THROUGHOUT / ESU R: 1.22 / ESU R Free: 0.39 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: BABINET MODEL WITH MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 31.306 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.7→65 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.7→2.77 Å / Total num. of bins used: 20 /
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller













 PDBj
PDBj