[English] 日本語
 Yorodumi
Yorodumi- PDB-1qoz: Catalytic core domain of acetyl xylan esterase from Trichoderma reesei -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1qoz | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Catalytic core domain of acetyl xylan esterase from Trichoderma reesei | |||||||||
 Components Components | ACETYL XYLAN ESTERASE | |||||||||
 Keywords Keywords |  HYDROLASE / HYDROLASE /  ESTERASE / XYLAN DEGRADATION ESTERASE / XYLAN DEGRADATION | |||||||||
| Function / homology |  Function and homology information Function and homology information acetylxylan esterase / acetylxylan esterase /  acetylxylan esterase activity / acetylxylan esterase activity /  cellulose binding / xylan catabolic process / cellulose catabolic process / extracellular region cellulose binding / xylan catabolic process / cellulose catabolic process / extracellular regionSimilarity search - Function | |||||||||
| Biological species |   TRICHODERMA REESEI (fungus) TRICHODERMA REESEI (fungus) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 1.9 Å MOLECULAR REPLACEMENT / Resolution: 1.9 Å | |||||||||
 Authors Authors | Hakulinen, N. / Rouvinen, J. | |||||||||
 Citation Citation |  Journal: J.Struct.Biol. / Year: 2000 Journal: J.Struct.Biol. / Year: 2000Title: Three-Dimensional Structure of the Catalytic Core of Acetylxylan Esterase from Trichoderma Reesei: Insights Into the Deacetylation Mechanism Authors: Hakulinen, N. / Tenkanen, M. / Rouvinen, J. #1: Journal: Acta Crystallogr.,Sect.D / Year: 1998 Title: Crystallization and Preliminary X-Ray Diffraction Studies of the Catalytic Core of Acetylxylan Esterase from Trichoderma Reesei Authors: Hakulinen, N. / Tenkanen, M. / Rouvinen, J. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1qoz.cif.gz 1qoz.cif.gz | 94.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1qoz.ent.gz pdb1qoz.ent.gz | 71.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1qoz.json.gz 1qoz.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/qo/1qoz https://data.pdbj.org/pub/pdb/validation_reports/qo/1qoz ftp://data.pdbj.org/pub/pdb/validation_reports/qo/1qoz ftp://data.pdbj.org/pub/pdb/validation_reports/qo/1qoz | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1bs9S S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 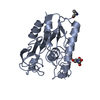
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
| ||||||||
| Noncrystallographic symmetry (NCS) | NCS oper: (Code: given Matrix: (-0.6846, 0.2911, -0.6682), Vector  : : |
- Components
Components
| #1: Protein | Mass: 21009.152 Da / Num. of mol.: 2 / Fragment: CATALYTIC DOMAIN, RESIDUES 32-238 / Source method: isolated from a natural source / Source: (natural)   TRICHODERMA REESEI (fungus) / Strain: RUTC-30 / References: UniProt: Q99034, TRICHODERMA REESEI (fungus) / Strain: RUTC-30 / References: UniProt: Q99034,  acetylxylan esterase acetylxylan esterase#2: Sugar |  N-Acetylglucosamine N-Acetylglucosamine#3: Water | ChemComp-HOH / |  Water Water |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.23 Å3/Da / Density % sol: 45 % | ||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
Crystal grow | pH: 8.2 Details: 1.1 M POTASSIUM/SODIUM TARTRATE, 0.1 M TES, 9 MM TRITON-X, PH 8.2 | ||||||||||||||||||||||||||||||||||||||||||||||||
| Crystal | *PLUS Density % sol: 45 % | ||||||||||||||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Method: vapor diffusion, hanging drop / pH: 8.2 | ||||||||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 120 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU RUH2R / Wavelength: 1.5418 ROTATING ANODE / Type: RIGAKU RUH2R / Wavelength: 1.5418 |
| Detector | Type: RIGAKU IMAGE PLATE / Detector: IMAGE PLATE / Date: Dec 15, 1998 |
| Radiation | Monochromator: GRAPHITE(002) / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 1.5418 Å / Relative weight: 1 : 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 1.9→99 Å / Num. obs: 27058 / % possible obs: 87 % / Redundancy: 2.07 % / Biso Wilson estimate: 18.76 Å2 / Rsym value: 0.057 / Net I/σ(I): 14.9 |
| Reflection shell | Resolution: 1.9→1.97 Å / Redundancy: 2 % / Mean I/σ(I) obs: 2.38 / Rsym value: 0.247 / % possible all: 46.5 |
| Reflection | *PLUS % possible obs: 87 % / Num. measured all: 55958 / Rmerge(I) obs: 0.057 |
| Reflection shell | *PLUS Highest resolution: 1.9 Å / % possible obs: 47 % / Rmerge(I) obs: 0.247 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1BS9 Resolution: 1.9→10 Å / Data cutoff high absF: 100000 / Data cutoff low absF: 0.1 / Cross valid method: THROUGHOUT / σ(F): 1
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 15.12 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.9→10 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.9→1.99 Å / Total num. of bins used: 8
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Xplor file | Serial no: 1 / Param file: PARHCSDX.PRO / Topol file: TOPHCSDX.PRO | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name:  X-PLOR / Version: 3.851 / Classification: refinement X-PLOR / Version: 3.851 / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | *PLUS Rfactor Rfree: 0.32 |
 Movie
Movie Controller
Controller











 PDBj
PDBj


