[English] 日本語
 Yorodumi
Yorodumi- PDB-1qjg: Crystal structure of delta5-3-ketosteroid isomerase from Pseudomo... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1qjg | ||||||
|---|---|---|---|---|---|---|---|
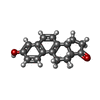
| Title | Crystal structure of delta5-3-ketosteroid isomerase from Pseudomonas testosteroni in complex with equilenin | ||||||
 Components Components | KETOSTEROID ISOMERASE | ||||||
 Keywords Keywords |  ISOMERASE / KETOSTEROID ISOMERASE / KSI / STEROID ISOMERATION ISOMERASE / KETOSTEROID ISOMERASE / KSI / STEROID ISOMERATION | ||||||
| Function / homology |  Function and homology information Function and homology information steroid Delta-isomerase / steroid Delta-isomerase /  steroid delta-isomerase activity / steroid metabolic process steroid delta-isomerase activity / steroid metabolic processSimilarity search - Function | ||||||
| Biological species |   PSEUDOMONAS TESTOSTERONI (bacteria) PSEUDOMONAS TESTOSTERONI (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 2.3 Å MOLECULAR REPLACEMENT / Resolution: 2.3 Å | ||||||
 Authors Authors | Cho, H.-S. / Oh, B.-H. | ||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 1999 Journal: J.Biol.Chem. / Year: 1999Title: Crystal Structure of Delta-5-3-Ketosteroid Isomerase from Pseudomonas Testosteroni in Complex with Equilenin Settles the Correct Hydrogen Scheme for Transition-State Stabilization Authors: Cho, H.-S. / Ha, N.-C. / Choi, G. / Kim, H.-J. / Lee, D. / Oh, K.S. / Kim, K.S. / Lee, W. / Choi, K.Y. / Oh, B.-H. #1:  Journal: Biochemistry / Year: 1998 Journal: Biochemistry / Year: 1998Title: Crystal Structure of Delta-Ketosteroid Isomerase from Pseudomonas Testosteroni. Authors: Cho, H.-S. / Choi, G. / Choi, K.Y. / Oh, B.-H. | ||||||
| History |
| ||||||
| Remark 650 | HELIX DETERMINATION METHOD: AUTHOR PROVIDED. | ||||||
| Remark 700 | SHEET DETERMINATION METHOD: AUTHOR PROVIDED. |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1qjg.cif.gz 1qjg.cif.gz | 187.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1qjg.ent.gz pdb1qjg.ent.gz | 152.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1qjg.json.gz 1qjg.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/qj/1qjg https://data.pdbj.org/pub/pdb/validation_reports/qj/1qjg ftp://data.pdbj.org/pub/pdb/validation_reports/qj/1qjg ftp://data.pdbj.org/pub/pdb/validation_reports/qj/1qjg | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  8choS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| 3 | 
| ||||||||
| Unit cell |
| ||||||||
| Details | THE ASYMMETRIC UNIT CONTAINS 3 INDEPENDENT COPIES OF THEBIOMOLECULE WHICH CONSISTS OF A HOMO -DIMERIC-COMPLEX |
- Components
Components
| #1: Protein | Mass: 13423.171 Da / Num. of mol.: 6 / Mutation: YES Source method: isolated from a genetically manipulated source Details: EQUILENIN, SO4 / Source: (gene. exp.)   PSEUDOMONAS TESTOSTERONI (bacteria) / Production host: PSEUDOMONAS TESTOSTERONI (bacteria) / Production host:   ESCHERICHIA COLI (E. coli) / Strain (production host): BL21(DE3) / References: UniProt: P00947 ESCHERICHIA COLI (E. coli) / Strain (production host): BL21(DE3) / References: UniProt: P00947#2: Chemical | ChemComp-EQU /  Equilenin Equilenin#3: Chemical | ChemComp-SO4 / |  Sulfate Sulfate#4: Water | ChemComp-HOH / |  Water Water |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.5 Å3/Da / Density % sol: 60 % | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
Crystal grow | pH: 4.6 Details: PEG 4000 25%, 0.1M SODIUM ACETATE, PH 4.6, 0.2M AMMONIUM SULFATE | |||||||||||||||||||||||||
| Crystal | *PLUS | |||||||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 12 ℃ / Method: vapor diffusion | |||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 287 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Wavelength: 1.5418 ROTATING ANODE / Wavelength: 1.5418 |
| Detector | Type: MACSCIENCE / Detector: IMAGE PLATE |
| Radiation | Monochromator: GRAPHITE(002) / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 1.5418 Å / Relative weight: 1 : 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 2.26→20 Å / Num. obs: 37500 / % possible obs: 96 % / Observed criterion σ(I): 1 / Redundancy: 2 % / Rmerge(I) obs: 0.05 / Rsym value: 0.069 |
| Reflection shell | Resolution: 2.26→2.34 Å / % possible all: 90 |
| Reflection | *PLUS Num. measured all: 102917 / Rmerge(I) obs: 0.069 |
| Reflection shell | *PLUS % possible obs: 90 % |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 8CHO Resolution: 2.3→10 Å / Cross valid method: THROUGHOUT / σ(F): 1
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.3→10 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name:  X-PLOR / Version: 3 / Classification: refinement X-PLOR / Version: 3 / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Rfactor obs: 0.196 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller













 PDBj
PDBj