[English] 日本語
 Yorodumi
Yorodumi- PDB-1qad: Crystal Structure of the C-Terminal SH2 Domain of the P85 alpha R... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1qad | ||||||
|---|---|---|---|---|---|---|---|
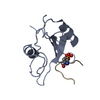
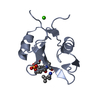
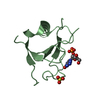
| Title | Crystal Structure of the C-Terminal SH2 Domain of the P85 alpha Regulatory Subunit of Phosphoinositide 3-Kinase: An SH2 domain mimicking its own substrate | ||||||
 Components Components | PI3-KINASE P85 ALPHA SUBUNIT | ||||||
 Keywords Keywords |  TRANSFERASE / PHOPHOTYROSINE-BINDING DOMAIN / SUBSTRATE MIMICKING TRANSFERASE / PHOPHOTYROSINE-BINDING DOMAIN / SUBSTRATE MIMICKING | ||||||
| Function / homology |  Function and homology information Function and homology informationSignaling by ALK / MET activates PI3K/AKT signaling / RHOC GTPase cycle / CDC42 GTPase cycle / RAC1 GTPase cycle / RAC2 GTPase cycle / RHOD GTPase cycle / RHOJ GTPase cycle / RAC3 GTPase cycle / RHOF GTPase cycle ...Signaling by ALK / MET activates PI3K/AKT signaling / RHOC GTPase cycle / CDC42 GTPase cycle / RAC1 GTPase cycle / RAC2 GTPase cycle / RHOD GTPase cycle / RHOJ GTPase cycle / RAC3 GTPase cycle / RHOF GTPase cycle / FLT3 Signaling / RND3 GTPase cycle / RND2 GTPase cycle / RND1 GTPase cycle / PI3K events in ERBB4 signaling / Interleukin-7 signaling / GAB1 signalosome / PI3K events in ERBB2 signaling / Erythropoietin activates Phosphoinositide-3-kinase (PI3K) / IRS-mediated signalling / GPVI-mediated activation cascade / Signaling by SCF-KIT / Downstream signal transduction / PI3K/AKT activation / Role of phospholipids in phagocytosis / Tie2 Signaling / Role of LAT2/NTAL/LAB on calcium mobilization / Costimulation by the CD28 family / CD28 dependent PI3K/Akt signaling / RAF/MAP kinase cascade / Interleukin receptor SHC signaling / PI-3K cascade:FGFR1 / PI-3K cascade:FGFR2 / PI-3K cascade:FGFR3 / PI-3K cascade:FGFR4 / Antigen activates B Cell Receptor (BCR) leading to generation of second messengers / PI3K Cascade / PIP3 activates AKT signaling / GP1b-IX-V activation signalling / PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling / Synthesis of PIPs at the plasma membrane / RHOA GTPase cycle / DAP12 signaling / RHOU GTPase cycle / RHOV GTPase cycle / Regulation of signaling by CBL / Downstream TCR signaling / RHOG GTPase cycle / RET signaling /  Interleukin-3, Interleukin-5 and GM-CSF signaling / VEGFA-VEGFR2 Pathway / phosphatidylinositol 3-kinase regulator activity / phosphatidylinositol 3-kinase activator activity / 1-phosphatidylinositol-3-kinase regulator activity / ErbB-3 class receptor binding / Interleukin-3, Interleukin-5 and GM-CSF signaling / VEGFA-VEGFR2 Pathway / phosphatidylinositol 3-kinase regulator activity / phosphatidylinositol 3-kinase activator activity / 1-phosphatidylinositol-3-kinase regulator activity / ErbB-3 class receptor binding /  transmembrane receptor protein tyrosine kinase adaptor activity / positive regulation of endoplasmic reticulum unfolded protein response / enzyme-substrate adaptor activity / phosphatidylinositol 3-kinase complex, class IA / transmembrane receptor protein tyrosine kinase adaptor activity / positive regulation of endoplasmic reticulum unfolded protein response / enzyme-substrate adaptor activity / phosphatidylinositol 3-kinase complex, class IA /  phosphatidylinositol 3-kinase complex / Extra-nuclear estrogen signaling / G alpha (q) signalling events / intracellular glucose homeostasis / phosphatidylinositol phosphate biosynthetic process / phosphatidylinositol 3-kinase complex / Extra-nuclear estrogen signaling / G alpha (q) signalling events / intracellular glucose homeostasis / phosphatidylinositol phosphate biosynthetic process /  insulin receptor substrate binding / insulin receptor substrate binding /  phosphatidylinositol 3-kinase binding / phosphatidylinositol 3-kinase binding /  insulin-like growth factor receptor binding / response to endoplasmic reticulum stress / substrate adhesion-dependent cell spreading / positive regulation of RNA splicing / insulin-like growth factor receptor signaling pathway / phosphatidylinositol 3-kinase/protein kinase B signal transduction / positive regulation of glucose import / positive regulation of protein localization to plasma membrane / insulin-like growth factor receptor binding / response to endoplasmic reticulum stress / substrate adhesion-dependent cell spreading / positive regulation of RNA splicing / insulin-like growth factor receptor signaling pathway / phosphatidylinositol 3-kinase/protein kinase B signal transduction / positive regulation of glucose import / positive regulation of protein localization to plasma membrane /  insulin receptor binding / positive regulation of protein import into nucleus / cellular response to insulin stimulus / insulin receptor binding / positive regulation of protein import into nucleus / cellular response to insulin stimulus /  protein transport / insulin receptor signaling pathway / protein stabilization / negative regulation of apoptotic process / positive regulation of transcription by RNA polymerase II / identical protein binding / protein transport / insulin receptor signaling pathway / protein stabilization / negative regulation of apoptotic process / positive regulation of transcription by RNA polymerase II / identical protein binding /  nucleus nucleusSimilarity search - Function | ||||||
| Biological species |   Bos taurus (cattle) Bos taurus (cattle) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / Resolution: 1.8 Å SYNCHROTRON / Resolution: 1.8 Å | ||||||
 Authors Authors | Hoedemaeker, P.J. / Siegal, G. / Roe, M. / Driscoll, P.C. / Abrahams, J.P.A. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 1999 Journal: J.Mol.Biol. / Year: 1999Title: Crystal structure of the C-terminal SH2 domain of the p85alpha regulatory subunit of phosphoinositide 3-kinase: an SH2 domain mimicking its own substrate. Authors: Hoedemaeker, F.J. / Siegal, G. / Roe, S.M. / Driscoll, P.C. / Abrahams, J.P. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1qad.cif.gz 1qad.cif.gz | 60.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1qad.ent.gz pdb1qad.ent.gz | 44.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1qad.json.gz 1qad.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/qa/1qad https://data.pdbj.org/pub/pdb/validation_reports/qa/1qad ftp://data.pdbj.org/pub/pdb/validation_reports/qa/1qad ftp://data.pdbj.org/pub/pdb/validation_reports/qa/1qad | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1bfjS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 12615.052 Da / Num. of mol.: 1 / Fragment: C-TERMINAL SH2 DOMAIN Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Bos taurus (cattle) / References: UniProt: P23727 Bos taurus (cattle) / References: UniProt: P23727 |
|---|---|
| #2: Water | ChemComp-HOH /  Water Water |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.427 Å3/Da / Density % sol: 49.3 % | |||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
Crystal grow | Temperature: 277 K / Method: spontaneous crystallisation / pH: 7.5 Details: TRIS-HCL, NACL, EDTA, DTT, pH 7.5, SPONTANEOUS CRYSTALLISATION, temperature 277.0K | |||||||||||||||||||||||||||||||||||
| Components of the solutions |
| |||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Method: vapor diffusion, hanging drop / Details: Eck, M.J., (1993) Nature, 362, 87. / pH: 4.6 | |||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SRS SRS  / Beamline: PX7.2 / Wavelength: 1.488 / Beamline: PX7.2 / Wavelength: 1.488 |
| Detector | Type: MARRESEARCH / Detector: IMAGE PLATE / Date: Jun 5, 1998 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 1.488 Å / Relative weight: 1 : 1.488 Å / Relative weight: 1 |
| Reflection | Resolution: 1.8→23.44 Å / Num. all: 10947 / Num. obs: 10947 / % possible obs: 98.7 % / Redundancy: 3.4 % / Biso Wilson estimate: 18 Å2 / Net I/σ(I): 7.2 |
| Reflection shell | Resolution: 1.8→1.85 Å / Redundancy: 3 % / Mean I/σ(I) obs: 10.2 / % possible all: 84.5 |
| Reflection | *PLUS Num. measured all: 37237 / Rmerge(I) obs: 0.044 |
| Reflection shell | *PLUS % possible obs: 84.5 % / Num. unique obs: 677 / Num. measured obs: 2058 / Rmerge(I) obs: 0.068 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Starting model: 1BFJ Resolution: 1.8→23.44 Å / SU B: 2.3668 / SU ML: 0.07628 / Cross valid method: Rfree / ESU R Free: 0.14209 / Stereochemistry target values: ENGH & HUBER Details: CONJUGATED DIRECTION METHOD WITH MAXIMUM LIKELYHOOD TARGET (REFMAC_aniso)
| |||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.8→23.44 Å
| |||||||||||||||||||||||||
| Software | *PLUS Name: REFMAC / Classification: refinement | |||||||||||||||||||||||||
| Refinement | *PLUS Rfactor obs: 0.158 | |||||||||||||||||||||||||
| Solvent computation | *PLUS | |||||||||||||||||||||||||
| Displacement parameters | *PLUS | |||||||||||||||||||||||||
| Refine LS restraints | *PLUS
| |||||||||||||||||||||||||
| LS refinement shell | *PLUS Highest resolution: 1.8 Å / Lowest resolution: 1.85 Å / Rfactor Rfree: 0.231 / Num. reflection Rfree: 135 / Num. reflection Rwork: 1159 / Rfactor obs: 0.118 |
 Movie
Movie Controller
Controller









 PDBj
PDBj



