[English] 日本語
 Yorodumi
Yorodumi- PDB-1mk6: SOLUTION STRUCTURE OF THE 8,9-DIHYDRO-8-(N7-GUANYL)-9-HYDROXY-AFL... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1mk6 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SOLUTION STRUCTURE OF THE 8,9-DIHYDRO-8-(N7-GUANYL)-9-HYDROXY-AFLATOXIN B1 ADDUCT MISPAIRED WITH DEOXYADENOSINE | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords |  DNA / AFLATOXIN B1- Guanine Adduct Opposite an Adenine / mimicking GA Transition DNA / AFLATOXIN B1- Guanine Adduct Opposite an Adenine / mimicking GA Transition | |||||||||
| Function / homology | 8,9-DIHYDRO-9-HYDROXY-AFLATOXIN B1 /  DNA DNA Function and homology information Function and homology information | |||||||||
| Biological species | synthetic construct (others) | |||||||||
| Method |  SOLUTION NMR / distance geometry : MardiGras; simulated annealing, molecular dynamics : XPLOR; Average structure Calculation Addition of Solvent, and Counterions : AMBER; simulated annealing, Molecular Dynamics matrix relaxation : CORMA; SOLUTION NMR / distance geometry : MardiGras; simulated annealing, molecular dynamics : XPLOR; Average structure Calculation Addition of Solvent, and Counterions : AMBER; simulated annealing, Molecular Dynamics matrix relaxation : CORMA; | |||||||||
 Authors Authors | Giri, I. / Johnston, D.S. / Stone, M.P. | |||||||||
 Citation Citation |  Journal: Biochemistry / Year: 2002 Journal: Biochemistry / Year: 2002Title: MISPAIRING OF THE 8,9-DIHYDRO-8-(N7-GUANYL)-9-HYDROXY-AFLATOXIN B1 ADDUCT WITH DEOXYADENOSINE RESULTS IN EXTRUSION OF THE MISMATCHED DA TOWARD THE MAJOR GROOVE Authors: Giri, I. / Johnston, D.S. / Stone, M.P. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1mk6.cif.gz 1mk6.cif.gz | 24.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1mk6.ent.gz pdb1mk6.ent.gz | 15.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1mk6.json.gz 1mk6.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/mk/1mk6 https://data.pdbj.org/pub/pdb/validation_reports/mk/1mk6 ftp://data.pdbj.org/pub/pdb/validation_reports/mk/1mk6 ftp://data.pdbj.org/pub/pdb/validation_reports/mk/1mk6 | HTTPS FTP |
|---|
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 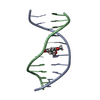
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: DNA chain | Mass: 3003.993 Da / Num. of mol.: 1 / Source method: obtained synthetically / Source: (synth.) synthetic construct (others) |
|---|---|
| #2: DNA chain | Mass: 3108.065 Da / Num. of mol.: 1 / Source method: obtained synthetically / Source: (synth.) synthetic construct (others) |
| #3: Chemical | ChemComp-AFN / |
-Experimental details
-Experiment
| Experiment | Method:  SOLUTION NMR SOLUTION NMR | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||||||||||
| NMR details | Text: 40 Structures were calculated starting from B and A form DNA. 20 closely convergent from both was averaged, and RMSD value was checked. The final averaged structure was calculated by averaging ...Text: 40 Structures were calculated starting from B and A form DNA. 20 closely convergent from both was averaged, and RMSD value was checked. The final averaged structure was calculated by averaging Final A and Final B, and after energy Minimization. This energy minimized structure was solvated, and explicit counterions were added. In all, 17 Na+ ions were added to neutralize the system using the Leap module in AMBER 6.0. The SHAKE algorithm constrained bonds involving protons to a tolerance of 0.0005 . A 1 fs time step was used. The rMD calculations were run for 1.4 ns, and coordinates were captured every 200 ps. The emergent structure from AMBER is Being reported |
- Sample preparation
Sample preparation
| Details | Contents: 80 OD of d(ACATCAFBGATCT)d(AGATAGATGT) solution in NMR Buffer Solvent system: For observation of nonexchangeable protons, the sample was dissolved in 0.5 mL of 0.01 M sodium phosphate containing 0.1 M NaCl and 0.05 mM Na2EDTA at pD 7.4. The sample was dissolved ...Solvent system: For observation of nonexchangeable protons, the sample was dissolved in 0.5 mL of 0.01 M sodium phosphate containing 0.1 M NaCl and 0.05 mM Na2EDTA at pD 7.4. The sample was dissolved in 99.96% D2O. For observation of exchangeable protons, the sample was dissolved in 9:1 H2O:D2O. Most experiments were performed at 5 C. Spectra of exchangeable protons were obtained at 0 C. | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample conditions |
| |||||||||||||||
Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Radiation wavelength | Relative weight: 1 | ||||||||||||||||||||
| NMR spectrometer |
|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: distance geometry : MardiGras; simulated annealing, molecular dynamics : XPLOR; Average structure Calculation Addition of Solvent, and Counterions : AMBER; simulated annealing, Molecular ...Method: distance geometry : MardiGras; simulated annealing, molecular dynamics : XPLOR; Average structure Calculation Addition of Solvent, and Counterions : AMBER; simulated annealing, Molecular Dynamics matrix relaxation : CORMA; Software ordinal: 1 Details: There were 329 experimental distance restraints derived from nonexchangeable 1H NOEs by MARDIGRAS. These consisted of 181 intranucleotide restraints, 110 internucleotide restraints, and 38 adduct-DNA restraints | ||||||||||||||||||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: Final Calculated Structure is Being Submitted. Back Calculated Structure is in Agreement with NOESY data. The calculation was performed in Presence of solvent and ...Conformer selection criteria: Final Calculated Structure is Being Submitted. Back Calculated Structure is in Agreement with NOESY data. The calculation was performed in Presence of solvent and counterions. Solvent, and Counterion co-ordinates are NOT being reported, only the Duplex DNA. Before Solvating and Addition of Counter IONS, 20 final structures were calculated using XPLOR. The final averaged energy minimized structure was solvated, and the counter Ions were added to it. Then MD was ran for 1.4 ns time scale to obtain final structure. Conformers calculated total number: 20 / Conformers submitted total number: 1 |
 Movie
Movie Controller
Controller








 PDBj
PDBj



