[English] 日本語
 Yorodumi
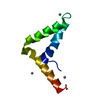
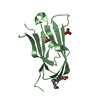
Yorodumi- PDB-1kg1: NMR structure of the NIP1 elicitor protein from Rhynchosporium secalis -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1kg1 | ||||||
|---|---|---|---|---|---|---|---|
| Title | NMR structure of the NIP1 elicitor protein from Rhynchosporium secalis | ||||||
 Components Components | Necrosis Inducing Protein 1 | ||||||
 Keywords Keywords |  TOXIN / antiparalel beta sheets TOXIN / antiparalel beta sheets | ||||||
| Function / homology |  Function and homology information Function and homology informationSH3 type barrels. - #460 /  Rhinovirus 14, subunit 4 - #10 / Necrosis inducing protein-1 / Necrosis inducing protein-1 superfamily / Necrosis inducing protein-1 / Rhinovirus 14, subunit 4 - #10 / Necrosis inducing protein-1 / Necrosis inducing protein-1 superfamily / Necrosis inducing protein-1 /  Rhinovirus 14, subunit 4 / Other non-globular / Special / SH3 type barrels. / Roll / Mainly Beta Rhinovirus 14, subunit 4 / Other non-globular / Special / SH3 type barrels. / Roll / Mainly BetaSimilarity search - Domain/homology | ||||||
| Biological species |   Rhynchosporium secalis (leaf blotch of barley) Rhynchosporium secalis (leaf blotch of barley) | ||||||
| Method |  SOLUTION NMR / torsion angle dynamics SOLUTION NMR / torsion angle dynamics | ||||||
 Authors Authors | Van't Slot, K.A. / Van den Burg, H.A. / Kloks, C.P. / Hilbers, C.W. / Knogge, W. / Papavoine, C.H. | ||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 2003 Journal: J.Biol.Chem. / Year: 2003Title: Solution Structure of the Plant Disease Resistance-triggering Protein NIP1 from the Fungus Rhynchosporium secalis Shows a Novel beta-Sheet Fold. Authors: Van't Slot, K.A. / Van Den Burg, H.A. / Kloks, C.P. / Hilbers, C.W. / Knogge, W. / Papavoine, C.H. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1kg1.cif.gz 1kg1.cif.gz | 408.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1kg1.ent.gz pdb1kg1.ent.gz | 353.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1kg1.json.gz 1kg1.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/kg/1kg1 https://data.pdbj.org/pub/pdb/validation_reports/kg/1kg1 ftp://data.pdbj.org/pub/pdb/validation_reports/kg/1kg1 ftp://data.pdbj.org/pub/pdb/validation_reports/kg/1kg1 | HTTPS FTP |
|---|
-Related structure data
| Similar structure data | |
|---|---|
| Other databases |
|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 6451.364 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Rhynchosporium secalis (leaf blotch of barley) Rhynchosporium secalis (leaf blotch of barley)Gene: Nip1 / Plasmid: pQE30 / Production host:   Escherichia coli (E. coli) / Strain (production host): AD494 / References: UniProt: Q02039 Escherichia coli (E. coli) / Strain (production host): AD494 / References: UniProt: Q02039 |
|---|
-Experimental details
-Experiment
| Experiment | Method:  SOLUTION NMR SOLUTION NMR | ||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||||||||||||||||||||||||||||||
| NMR details | Text: Disulfide bond pattern was determined independently |
- Sample preparation
Sample preparation
| Details |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample conditions | pH: 6.0 / Pressure: ambient / Temperature: 298 K | ||||||||||||
Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Radiation wavelength | Relative weight: 1 | ||||||||||||||||||||
| NMR spectrometer |
|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: torsion angle dynamics / Software ordinal: 1 Details: The structures are besed on a total of 785 restraints: 740 NOE restraints and 45, 40 torsion angle restraints and 5 disulfide bonds | ||||||||||||||||||||
| NMR representative | Selection criteria: closest to the average | ||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: target function / Conformers calculated total number: 100 / Conformers submitted total number: 25 |
 Movie
Movie Controller
Controller








 PDBj
PDBj