+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1fi6 | ||||||
|---|---|---|---|---|---|---|---|
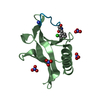
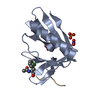
| Title | SOLUTION STRUCTURE OF THE REPS1 EH DOMAIN | ||||||
 Components Components | EH DOMAIN PROTEIN REPS1 EF hand EF hand | ||||||
 Keywords Keywords | endocytosis/exocytosis / EPS15 HOMOLOGY DOMAIN /  EF HAND / EF HAND /  CALCIUM / RAS SIGNAL TRANSDUCTION / endocytosis-exocytosis COMPLEX CALCIUM / RAS SIGNAL TRANSDUCTION / endocytosis-exocytosis COMPLEX | ||||||
| Function / homology |  Function and homology information Function and homology informationCargo recognition for clathrin-mediated endocytosis /  Clathrin-mediated endocytosis / endosomal transport / Clathrin-mediated endocytosis / endosomal transport /  clathrin-coated pit / clathrin-coated pit /  receptor-mediated endocytosis / receptor-mediated endocytosis /  SH3 domain binding / SH3 domain binding /  endocytosis / endocytosis /  calcium ion binding / calcium ion binding /  plasma membrane plasma membraneSimilarity search - Function | ||||||
| Biological species |   Mus musculus (house mouse) Mus musculus (house mouse) | ||||||
| Method |  SOLUTION NMR / SOLUTION NMR /  simulated annealing simulated annealing | ||||||
 Authors Authors | Kim, S. / Baleja, J.D. | ||||||
 Citation Citation |  Journal: Biochemistry / Year: 2001 Journal: Biochemistry / Year: 2001Title: Solution structure of the Reps1 EH domain and characterization of its binding to NPF target sequences. Authors: Kim, S. / Cullis, D.N. / Feig, L.A. / Baleja, J.D. #1:  Journal: Thesis / Year: 2000 Journal: Thesis / Year: 2000Title: Chapter 3: Studies of Protein Structure and Interaction using Nuclear Magnetic Resonance Spectroscopy:Applications to the Reps1 EH Domain and MicrocinB17 Propeptide Authors: Kim, S. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1fi6.cif.gz 1fi6.cif.gz | 868.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1fi6.ent.gz pdb1fi6.ent.gz | 724.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1fi6.json.gz 1fi6.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/fi/1fi6 https://data.pdbj.org/pub/pdb/validation_reports/fi/1fi6 ftp://data.pdbj.org/pub/pdb/validation_reports/fi/1fi6 ftp://data.pdbj.org/pub/pdb/validation_reports/fi/1fi6 | HTTPS FTP |
|---|
-Related structure data
| Similar structure data | |
|---|---|
| Other databases |
|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein |  EF hand EF handMass: 10647.128 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Mus musculus (house mouse) / Plasmid: PGEX2T / Production host: Mus musculus (house mouse) / Plasmid: PGEX2T / Production host:   Escherichia coli (E. coli) / References: UniProt: O54916 Escherichia coli (E. coli) / References: UniProt: O54916 |
|---|---|
| #2: Chemical | ChemComp-CA / |
-Experimental details
-Experiment
| Experiment | Method:  SOLUTION NMR SOLUTION NMR | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||||||
| NMR details | Text: The structure was determined using heteronuclear 2D and 3D NMR spectroscopy. |
- Sample preparation
Sample preparation
| Details |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample conditions | Ionic strength: 10mM NaCl / pH: 6.8 / Pressure: ambient / Temperature: 30 K | ||||||||||||
Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| NMR spectrometer |
|
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method:  simulated annealing / Software ordinal: 1 simulated annealing / Software ordinal: 1 Details: The structures are based on a total of 1265 restraints, 1143 are NOE-derived distance constraints, 122 dihedral angle restraints. | ||||||||||||||||||||||||
| NMR representative | Selection criteria: closest to the average | ||||||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the lowest energy Conformers calculated total number: 40 / Conformers submitted total number: 30 |
 Movie
Movie Controller
Controller











 PDBj
PDBj




