[English] 日本語
 Yorodumi
Yorodumi- EMDB-8944: 20S supercomplex consisting of linked neuronal SNARE complex, alp... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8944 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | 20S supercomplex consisting of linked neuronal SNARE complex, alpha-SNAP, and N-ethylmaleimide sensitive factor (NSF) | |||||||||
 Map data Map data | 20S supercomplex with linked SNARE | |||||||||
 Sample Sample |
| |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 7.0 Å cryo EM / Resolution: 7.0 Å | |||||||||
 Authors Authors | Zhao M / Choi UB / Brunger AT | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2018 Journal: Elife / Year: 2018Title: NSF-mediated disassembly of on- and off-pathway SNARE complexes and inhibition by complexin. Authors: Ucheor B Choi / Minglei Zhao / K Ian White / Richard A Pfuetzner / Luis Esquivies / Qiangjun Zhou / Axel T Brunger /  Abstract: SNARE complex disassembly by the ATPase NSF is essential for neurotransmitter release and other membrane trafficking processes. We developed a single-molecule FRET assay to monitor repeated rounds of ...SNARE complex disassembly by the ATPase NSF is essential for neurotransmitter release and other membrane trafficking processes. We developed a single-molecule FRET assay to monitor repeated rounds of NSF-mediated disassembly and reassembly of individual SNARE complexes. For ternary neuronal SNARE complexes, disassembly proceeds in a single step within 100 msec. We observed short- (<0.32 s) and long-lived (≥0.32 s) disassembled states. The long-lived states represent fully disassembled SNARE complex, while the short-lived states correspond to failed disassembly or immediate reassembly. Either high ionic strength or decreased αSNAP concentration reduces the disassembly rate while increasing the frequency of short-lived states. NSF is also capable of disassembling anti-parallel ternary SNARE complexes, implicating it in quality control. Finally, complexin-1 competes with αSNAP binding to the SNARE complex; addition of complexin-1 has an effect similar to that of decreasing the αSNAP concentration, possibly differentially regulating cis and trans SNARE complexes disassembly. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8944.map.gz emd_8944.map.gz | 9.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8944-v30.xml emd-8944-v30.xml emd-8944.xml emd-8944.xml | 11.8 KB 11.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8944.png emd_8944.png | 41.1 KB | ||
| Others |  emd_8944_additional.map.gz emd_8944_additional.map.gz | 12.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8944 http://ftp.pdbj.org/pub/emdb/structures/EMD-8944 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8944 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8944 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_8944.map.gz / Format: CCP4 / Size: 12.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8944.map.gz / Format: CCP4 / Size: 12.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 20S supercomplex with linked SNARE | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.02 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Sharpened map of 20S supercomplex with linked SNARE
| File | emd_8944_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map of 20S supercomplex with linked SNARE | ||||||||||||
| Projections & Slices |
| ||||||||||||
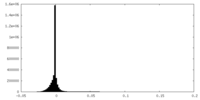
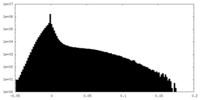
| Density Histograms |
- Sample components
Sample components
-Entire : 20S Supercomplex with linked SNARE complex
| Entire | Name: 20S Supercomplex with linked SNARE complex |
|---|---|
| Components |
|
-Supramolecule #1: 20S Supercomplex with linked SNARE complex
| Supramolecule | Name: 20S Supercomplex with linked SNARE complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 15 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Support film - Material: CARBON / Support film - topology: HOLEY / Details: unspecified |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm Bright-field microscopy / Cs: 2.7 mm |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 78.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Software - Name: Gctf |
|---|---|
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION (ver. 1.3) |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION (ver. 1.3) |
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 7.0 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 1.3) / Number images used: 45194 |
 Movie
Movie Controller
Controller











 Z
Z Y
Y X
X