[English] 日本語
 Yorodumi
Yorodumi- EMDB-8574: Single particle reconstruction of chimeric adeno-associated virus... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8574 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Single particle reconstruction of chimeric adeno-associated virus-DJ with a Heparanoid Pentasaccharide | |||||||||
 Map data Map data | chimeric adeno-associated virus-DJ with a Heparanoid Pentasaccharide | |||||||||
 Sample Sample |
| |||||||||
| Biological species |    Adeno-associated virus Adeno-associated virus | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.8 Å cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Xie Q / Noble AJ / Sousa DR / Meyer NL / Davulcu O / Zhang FM / Linhardt RJ / Stagg SM / Chapman MS | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Mol Ther Methods Clin Dev / Year: 2017 Journal: Mol Ther Methods Clin Dev / Year: 2017Title: The 2.8 Å Electron Microscopy Structure of Adeno-Associated Virus-DJ Bound by a Heparinoid Pentasaccharide. Authors: Qing Xie / John M Spear / Alex J Noble / Duncan R Sousa / Nancy L Meyer / Omar Davulcu / Fuming Zhang / Robert J Linhardt / Scott M Stagg / Michael S Chapman /  Abstract: Atomic structures of adeno-associated virus (AAV)-DJ, alone and in complex with fondaparinux, have been determined by cryoelectron microscopy at 3 Å resolution. The gene therapy vector, AAV-DJ, is ...Atomic structures of adeno-associated virus (AAV)-DJ, alone and in complex with fondaparinux, have been determined by cryoelectron microscopy at 3 Å resolution. The gene therapy vector, AAV-DJ, is a hybrid of natural serotypes that was previously derived by directed evolution, selecting for hepatocyte entry and resistance to neutralization by human serum. The structure of AAV-DJ differs from that of parental serotypes in two regions where neutralizing antibodies bind, so immune escape appears to have been the primary driver of AAV-DJ's directed evolution. Fondaparinux is an analog of cell surface heparan sulfate to which several AAVs bind during entry. Fondaparinux interacts with viral arginines at a known heparin binding site, without the large conformational changes whose presence was controversial in low-resolution imaging of AAV2-heparin complexes. The glycan density suggests multi-modal binding that could accommodate sequence variation and multivalent binding along a glycan polymer, consistent with a role in attachment, prior to more specific interactions with a receptor protein mediating entry. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8574.map.gz emd_8574.map.gz | 67.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8574-v30.xml emd-8574-v30.xml emd-8574.xml emd-8574.xml | 17.7 KB 17.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8574.png emd_8574.png | 628.7 KB | ||
| Others |  emd_8574_additional_1.map.gz emd_8574_additional_1.map.gz emd_8574_additional_2.map.gz emd_8574_additional_2.map.gz | 66.6 MB 68.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8574 http://ftp.pdbj.org/pub/emdb/structures/EMD-8574 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8574 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8574 | HTTPS FTP |
-Related structure data
| Related structure data |  5uf6MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_8574.map.gz / Format: CCP4 / Size: 76.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8574.map.gz / Format: CCP4 / Size: 76.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | chimeric adeno-associated virus-DJ with a Heparanoid Pentasaccharide | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.2159 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Adeno-Associated Virus-DJ Bound by a Heparanoid Pentasaccharide, native...
| File | emd_8574_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Adeno-Associated Virus-DJ Bound by a Heparanoid Pentasaccharide, native map | ||||||||||||
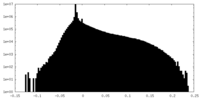
| Projections & Slices |
| ||||||||||||
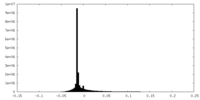
| Density Histograms |
-Additional map: Adeno-Associated Virus-DJ Bound by a Heparanoid Pentasaccharide, difference...
| File | emd_8574_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Adeno-Associated Virus-DJ Bound by a Heparanoid Pentasaccharide, difference map | ||||||||||||
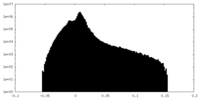
| Projections & Slices |
| ||||||||||||
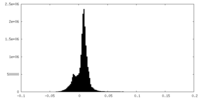
| Density Histograms |
- Sample components
Sample components
-Entire : Adeno-associated virus
| Entire | Name:    Adeno-associated virus Adeno-associated virus |
|---|---|
| Components |
|
-Supramolecule #1: Adeno-associated virus
| Supramolecule | Name: Adeno-associated virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1 / NCBI-ID: 272636 / Sci species name: Adeno-associated virus / Sci species strain: hybrid of serotypes 2, 8, and 9 / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: SEROTYPE / Virus enveloped: No / Virus empty: Yes |
|---|---|
| Host system | Organism:   Spodoptera frugiperda (fall armyworm) / Recombinant strain: SF9 / Recombinant plasmid: pFBDDJM11 Spodoptera frugiperda (fall armyworm) / Recombinant strain: SF9 / Recombinant plasmid: pFBDDJM11 |
| Molecular weight | Theoretical: 3.75 MDa |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.60 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||||||
| Grid | Model: Quantifoil / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Atmosphere: OTHER | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: blot force = 1, blot time = 3 seconds, total blots = 1. | ||||||||||||
| Details | 60 viral subunits form the icosahedral capsid |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.75 µm / Nominal magnification: 29000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.75 µm / Nominal magnification: 29000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: DIRECT ELECTRON DE-20 (5k x 3k) / Detector mode: INTEGRATING / Digitization - Frames/image: 1-45 / Number grids imaged: 1 / Number real images: 1051 / Average exposure time: 1.4 sec. / Average electron dose: 66.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Particle selection | Number selected: 120166 |
|---|---|
| CTF correction | Software: (Name: ACE, CTFFIND) |
| Initial angle assignment | Type: PROJECTION MATCHING / Software - Name: FREALIGN |
| Final angle assignment | Type: PROJECTION MATCHING / Software - Name: FREALIGN |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.8 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: FREALIGN / Number images used: 107454 |
 Movie
Movie Controller
Controller













 Z
Z Y
Y X
X