[English] 日本語
 Yorodumi
Yorodumi- EMDB-8313: Structure of the SLC4 transporter Bor1p in an inward-facing confo... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8313 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the SLC4 transporter Bor1p in an inward-facing conformation | |||||||||||||||
 Map data Map data | Bor1p helical tube density map (Type 1) | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | boron transporter /  anion exchanger family / alternating access mechanism / anion exchanger family / alternating access mechanism /  Structural Genomics / PSI-Biology / Transcontinental EM Initiative for Membrane Protein Structure / TEMIMPS / Structural Genomics / PSI-Biology / Transcontinental EM Initiative for Membrane Protein Structure / TEMIMPS /  TRANSPORT PROTEIN TRANSPORT PROTEIN | |||||||||||||||
| Function / homology |  Bicarbonate transporter, eukaryotic / Bicarbonate transporter-like, transmembrane domain / HCO3- transporter family / solute:inorganic anion antiporter activity / membrane => GO:0016020 / Bor1p boron transporter Bicarbonate transporter, eukaryotic / Bicarbonate transporter-like, transmembrane domain / HCO3- transporter family / solute:inorganic anion antiporter activity / membrane => GO:0016020 / Bor1p boron transporter Function and homology information Function and homology information | |||||||||||||||
| Biological species |   Saccharomyces mikatae (yeast) Saccharomyces mikatae (yeast) | |||||||||||||||
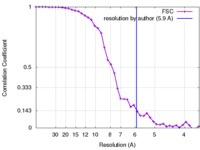
| Method | helical reconstruction /  cryo EM / Resolution: 5.9 Å cryo EM / Resolution: 5.9 Å | |||||||||||||||
 Authors Authors | Coudray N / Seyler S | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Protein Sci / Year: 2017 Journal: Protein Sci / Year: 2017Title: Structure of the SLC4 transporter Bor1p in an inward-facing conformation. Authors: Nicolas Coudray / Sean L Seyler / Ralph Lasala / Zhening Zhang / Kathy M Clark / Mark E Dumont / Alexis Rohou / Oliver Beckstein / David L Stokes /  Abstract: Bor1p is a secondary transporter in yeast that is responsible for boron transport. Bor1p belongs to the SLC4 family which controls bicarbonate exchange and pH regulation in animals as well as borate ...Bor1p is a secondary transporter in yeast that is responsible for boron transport. Bor1p belongs to the SLC4 family which controls bicarbonate exchange and pH regulation in animals as well as borate uptake in plants. The SLC4 family is more distantly related to members of the Amino acid-Polyamine-organoCation (APC) superfamily, which includes well studied transporters such as LeuT, Mhp1, AdiC, vSGLT, UraA, SLC26Dg. Their mechanism generally involves relative movements of two domains: a core domain that binds substrate and a gate domain that in many cases mediates dimerization. To shed light on conformational changes governing transport by the SLC4 family, we grew helical membrane crystals of Bor1p from Saccharomyces mikatae and determined a structure at ∼6 Å resolution using cryo-electron microscopy. To evaluate the conformation of Bor1p in these crystals, a homology model was built based on the related anion exchanger from red blood cells (AE1). This homology model was fitted to the cryo-EM density map using the Molecular Dynamics (MD) Flexible Fitting method and then relaxed by all-atom MD simulation in explicit solvent and membrane. Mapping of water accessibility indicates that the resulting structure represents an inward-facing conformation. Comparisons of the resulting Bor1p model with the X-ray structure of AE1 in an outward-facing conformation, together with MD simulations of inward-facing and outward-facing Bor1p models, suggest rigid body movements of the core domain relative to the gate domain. These movements are consistent with the rocking-bundle transport mechanism described for other members of the APC superfamily. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8313.map.gz emd_8313.map.gz | 500 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8313-v30.xml emd-8313-v30.xml emd-8313.xml emd-8313.xml | 17.9 KB 17.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_8313_fsc.xml emd_8313_fsc.xml | 4.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_8313.png emd_8313.png | 266.1 KB | ||
| Filedesc metadata |  emd-8313.cif.gz emd-8313.cif.gz | 6.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8313 http://ftp.pdbj.org/pub/emdb/structures/EMD-8313 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8313 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8313 | HTTPS FTP |
-Related structure data
| Related structure data |  5sv9MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_8313.map.gz / Format: CCP4 / Size: 3.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8313.map.gz / Format: CCP4 / Size: 3.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Bor1p helical tube density map (Type 1) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.82 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Bor1p dimer in an inward-facing conformation
| Entire | Name: Bor1p dimer in an inward-facing conformation |
|---|---|
| Components |
|
-Supramolecule #1: Bor1p dimer in an inward-facing conformation
| Supramolecule | Name: Bor1p dimer in an inward-facing conformation / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Saccharomyces mikatae (yeast) Saccharomyces mikatae (yeast) |
| Molecular weight | Theoretical: 60 KDa |
-Macromolecule #1: Bor1p boron transporter
| Macromolecule | Name: Bor1p boron transporter / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Saccharomyces mikatae (yeast) Saccharomyces mikatae (yeast) |
| Molecular weight | Theoretical: 53.829766 KDa |
| Recombinant expression | Organism:   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast) |
| Sequence | String: IWLDLKDRIP YYKSDWVDAF NYRVIPSTVD TYFNNLLPAI AFAQDMFDRT DNSYGVNEVL LSSAMAGIVF GVLAGQPLCI VGVTGPISI FNYTVYEIIK PLNTSYFGFM FWICLWSMIF HLLLAFTNVV CLLQYVTTFP CDIFGLFINV VYIQKGIQIL T RQFHNTSG ...String: IWLDLKDRIP YYKSDWVDAF NYRVIPSTVD TYFNNLLPAI AFAQDMFDRT DNSYGVNEVL LSSAMAGIVF GVLAGQPLCI VGVTGPISI FNYTVYEIIK PLNTSYFGFM FWICLWSMIF HLLLAFTNVV CLLQYVTTFP CDIFGLFINV VYIQKGIQIL T RQFHNTSG EKSVQDGFAS VVVALVMTAF GLFFKSFHHY PLFTHKIRTF ISDYSTALSV LFWSSFTHFG GYLNDVKFKK LP ITKSFFP TSKFNRPQNT WLAYEPIPVK DVFIALPFGI ILTILFYFDH NVSSLMAQRH QYKLRKPSSF HYDFALLGLT TCI SGVLGI PAPNGLIPQA PLHTETLLVR DSNQNVVRCV EQRLTNTFQG LMILGTMTRP LLVCLGEIPQ AVLSGLFFIM GING LMTNV IIHRIVFLFS DPKRRDNNSP LAKISKRSMV IFLCFSLAGF TGEFAITNTI AAIGFPLVLL LSVIVSFSFT Y |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Concentration | 0.25 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7 Component:
Details: Buffer was changed twice per day. | |||||||||||||||
| Grid | Model: EMS / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: OTHER | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Instrument: HOMEMADE PLUNGER | |||||||||||||||
| Details | The protein was solubilized in 1% n-Dodecyl beta-D-maltoside and exchanged into heptaethyleneglycol-n-dodecylether (C12E7) while bound to the IgG Sepharose affinity column. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 2.7 µm / Calibrated defocus min: 0.85 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.0 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 19000 Bright-field microscopy / Cs: 2.0 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 19000 |
| Sample stage | Specimen holder model: GATAN 626 SINGLE TILT LIQUID NITROGEN CRYO TRANSFER HOLDER Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 2-27 / Number grids imaged: 2 / Number real images: 252 / Average exposure time: 6.75 sec. / Average electron dose: 45.0 e/Å2 |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Details | A homology model was first created using human AE1 (PDB entry 4YZF) as a template using MODELLER, placed into the density map using SITUS, and then fitted into the map using the Molecular Dynamics Flexible Fitting (MDFF) method. | ||||||
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Target criteria: cross-correlation coefficient | ||||||
| Output model |  PDB-5sv9: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)