[English] 日本語
 Yorodumi
Yorodumi- EMDB-6717: Contactin-2 reconstructed by Individual-Particle Electron Tomography -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6717 | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Contactin-2 reconstructed by Individual-Particle Electron Tomography | |||||||||||||||||||||
 Map data Map data | Contactin-2 conformation 1 | |||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||||||||||||||
| Method |  electron tomography / electron tomography /  negative staining / Resolution: 20.0 Å negative staining / Resolution: 20.0 Å | |||||||||||||||||||||
 Authors Authors | Lu Z / Liu JK / Ren G / Rudenko G | |||||||||||||||||||||
| Funding support |  United States, United States,  China, 6 items China, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2016 Journal: J Biol Chem / Year: 2016Title: Molecular Architecture of Contactin-associated Protein-like 2 (CNTNAP2) and Its Interaction with Contactin 2 (CNTN2). Authors: Zhuoyang Lu / M V V V Sekhar Reddy / Jianfang Liu / Ana Kalichava / Jiankang Liu / Lei Zhang / Fang Chen / Yun Wang / Luis Marcelo F Holthauzen / Mark A White / Suchithra Seshadrinathan / ...Authors: Zhuoyang Lu / M V V V Sekhar Reddy / Jianfang Liu / Ana Kalichava / Jiankang Liu / Lei Zhang / Fang Chen / Yun Wang / Luis Marcelo F Holthauzen / Mark A White / Suchithra Seshadrinathan / Xiaoying Zhong / Gang Ren / Gabby Rudenko /   Abstract: Contactin-associated protein-like 2 (CNTNAP2) is a large multidomain neuronal adhesion molecule implicated in a number of neurological disorders, including epilepsy, schizophrenia, autism spectrum ...Contactin-associated protein-like 2 (CNTNAP2) is a large multidomain neuronal adhesion molecule implicated in a number of neurological disorders, including epilepsy, schizophrenia, autism spectrum disorder, intellectual disability, and language delay. We reveal here by electron microscopy that the architecture of CNTNAP2 is composed of a large, medium, and small lobe that flex with respect to each other. Using epitope labeling and fragments, we assign the F58C, L1, and L2 domains to the large lobe, the FBG and L3 domains to the middle lobe, and the L4 domain to the small lobe of the CNTNAP2 molecular envelope. Our data reveal that CNTNAP2 has a very different architecture compared with neurexin 1α, a fellow member of the neurexin superfamily and a prototype, suggesting that CNTNAP2 uses a different strategy to integrate into the synaptic protein network. We show that the ectodomains of CNTNAP2 and contactin 2 (CNTN2) bind directly and specifically, with low nanomolar affinity. We show further that mutations in CNTNAP2 implicated in autism spectrum disorder are not segregated but are distributed over the whole ectodomain. The molecular shape and dimensions of CNTNAP2 place constraints on how CNTNAP2 integrates in the cleft of axo-glial and neuronal contact sites and how it functions as an organizing and adhesive molecule. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
-Related structure data
| Related structure data |  9543C  9544C  9545C 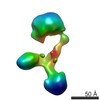 9546C  9547C  9548C  9549C  9550C  9551C  9552C  9553C  9554C  9555C 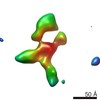 9556C  9557C 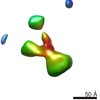 9558C  9559C  9560C  9561C  9562C  9563C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_6717.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6717.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Contactin-2 conformation 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.96 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
+Additional map: Contactin-2 conformation 2
+Additional map: Contactin-2 conformation 11
+Additional map: Contactin-2 conformation 12
+Additional map: Contactin-2 conformation 13
+Additional map: Contactin-2 conformation 14
+Additional map: Contactin-2 conformation 15
+Additional map: Contactin-2 conformation 16
+Additional map: Contactin-2 conformation 17
+Additional map: Contactin-2 conformation 18
+Additional map: Contactin-2 conformation 19
+Additional map: Contactin-2 conformation 20
+Additional map: Contactin-2 conformation 3
+Additional map: Contactin-2 conformation 21
+Additional map: Contactin-2 conformation 22
+Additional map: Contactin-2 conformation 23
+Additional map: Contactin-2 conformation 24
+Additional map: Contactin-2 conformation 25
+Additional map: Contactin-2 conformation 26
+Additional map: Contactin-2 conformation 27
+Additional map: Contactin-2 conformation 28
+Additional map: Contactin-2 conformation 29
+Additional map: Contactin-2 conformation 30
+Additional map: Contactin-2 conformation 4
+Additional map: Contactin-2 conformation 31
+Additional map: Contactin-2 conformation 32
+Additional map: Contactin-2 conformation 33
+Additional map: Contactin-2 conformation 34
+Additional map: Contactin-2 conformation 35
+Additional map: Contactin-2 conformation 36
+Additional map: Contactin-2 conformation 37
+Additional map: Contactin-2 conformation 38
+Additional map: Contactin-2 conformation 39
+Additional map: Contactin-2 conformation 40
+Additional map: Contactin-2 conformation 5
+Additional map: Contactin-2 conformation 41
+Additional map: Contactin-2 conformation 42
+Additional map: Contactin-2 conformation 43
+Additional map: Contactin-2 conformation 44
+Additional map: Contactin-2 conformation 45
+Additional map: Contactin-2 conformation 46
+Additional map: Contactin-2 conformation 47
+Additional map: Contactin-2 conformation 48
+Additional map: Contactin-2 conformation 49
+Additional map: Contactin-2 conformation 50
+Additional map: Contactin-2 conformation 6
+Additional map: Contactin-2 conformation 51
+Additional map: Contactin-2 conformation 52
+Additional map: Contactin-2 conformation 53
+Additional map: Contactin-2 conformation 54
+Additional map: Contactin-2 conformation 55
+Additional map: Contactin-2 conformation 56
+Additional map: Contactin-2 conformation 57
+Additional map: Contactin-2 conformation 58
+Additional map: Contactin-2 conformation 59
+Additional map: Contactin-2 conformation 7
+Additional map: Contactin-2 conformation 8
+Additional map: Contactin-2 conformation 9
+Additional map: Contactin-2 conformation 10
- Sample components
Sample components
-Entire : Contactin-2
| Entire | Name: Contactin-2 |
|---|---|
| Components |
|
-Supramolecule #1: Contactin-2
| Supramolecule | Name: Contactin-2 / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method |  negative staining negative staining |
|---|---|
 Processing Processing |  electron tomography electron tomography |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.0025 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||
| Staining | Type: NEGATIVE / Material: uranium formate / Details: 1% w/v uranyl formate | ||||||||||||
| Grid | Model: EMSCF, 200-Cu / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 5.0 nm / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: OTHER | ||||||||||||
| Details | This sample was monodisperse. | ||||||||||||
| Sectioning | Other: NO SECTIONING |
- Electron microscopy
Electron microscopy
| Microscope | ZEISS LIBRA120PLUS |
|---|---|
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.2 mm / Nominal defocus max: 0.6 µm / Nominal defocus min: 0.1 µm / Nominal magnification: 80000 Bright-field microscopy / Cs: 2.2 mm / Nominal defocus max: 0.6 µm / Nominal defocus min: 0.1 µm / Nominal magnification: 80000 |
| Sample stage | Specimen holder model: GATAN 626 SINGLE TILT LIQUID NITROGEN CRYO TRANSFER HOLDER |
| Image recording | Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Average electron dose: 3.0 e/Å2 |
- Image processing
Image processing
| CTF correction | Software - Name: TOMOCTF |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 20.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: IPET / Number images used: 81 |
 Movie
Movie Controller
Controller





 Z
Z Y
Y X
X