+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6655 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | 3DEM structure of Rrp5 bound to H45-3ITS1 RNA | |||||||||
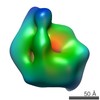
 Map data Map data | Reconstruction of Rrp5 bound to RNA | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Rrp5 /  RNA binding protein / ribosome assembly factor / 40S ribosome / 60S ribosome / Rok1 / DEAD-box protein RNA binding protein / ribosome assembly factor / 40S ribosome / 60S ribosome / Rok1 / DEAD-box protein | |||||||||
| Function / homology |  Function and homology information Function and homology informationintracellular anatomical structure /  protein binding / box H/ACA snoRNA binding / endonucleolytic cleavage in ITS1 upstream of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / box C/D sno(s)RNA binding / endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / rRNA primary transcript binding / 90S preribosome assembly / poly(U) RNA binding ...intracellular anatomical structure / protein binding / box H/ACA snoRNA binding / endonucleolytic cleavage in ITS1 upstream of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / box C/D sno(s)RNA binding / endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / rRNA primary transcript binding / 90S preribosome assembly / poly(U) RNA binding ...intracellular anatomical structure /  protein binding / box H/ACA snoRNA binding / endonucleolytic cleavage in ITS1 upstream of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / box C/D sno(s)RNA binding / endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / rRNA primary transcript binding / 90S preribosome assembly / poly(U) RNA binding / U3 snoRNA binding / protein binding / box H/ACA snoRNA binding / endonucleolytic cleavage in ITS1 upstream of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / box C/D sno(s)RNA binding / endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / rRNA primary transcript binding / 90S preribosome assembly / poly(U) RNA binding / U3 snoRNA binding /  snoRNA binding / Major pathway of rRNA processing in the nucleolus and cytosol / 90S preribosome / endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / snoRNA binding / Major pathway of rRNA processing in the nucleolus and cytosol / 90S preribosome / endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) /  RNA processing / maturation of SSU-rRNA / small-subunit processome / rRNA processing / RNA processing / maturation of SSU-rRNA / small-subunit processome / rRNA processing /  mRNA binding / mRNA binding /  nucleolus / nucleolus /  RNA binding / RNA binding /  nucleoplasm nucleoplasmSimilarity search - Function | |||||||||
| Biological species |   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  negative staining / Resolution: 30.0 Å negative staining / Resolution: 30.0 Å | |||||||||
 Authors Authors | Khoshnevis S / Askenasy I / Johnson MC / Young-Erdos CL / Stroupe ME / Karbstein K | |||||||||
 Citation Citation |  Journal: PLoS Biol / Year: 2016 Journal: PLoS Biol / Year: 2016Title: The DEAD-box Protein Rok1 Orchestrates 40S and 60S Ribosome Assembly by Promoting the Release of Rrp5 from Pre-40S Ribosomes to Allow for 60S Maturation. Authors: Sohail Khoshnevis / Isabel Askenasy / Matthew C Johnson / Maria D Dattolo / Crystal L Young-Erdos / M Elizabeth Stroupe / Katrin Karbstein /  Abstract: DEAD-box proteins are ubiquitous regulators of RNA biology. While commonly dubbed "helicases," their activities also include duplex annealing, adenosine triphosphate (ATP)-dependent RNA binding, and ...DEAD-box proteins are ubiquitous regulators of RNA biology. While commonly dubbed "helicases," their activities also include duplex annealing, adenosine triphosphate (ATP)-dependent RNA binding, and RNA-protein complex remodeling. Rok1, an essential DEAD-box protein, and its cofactor Rrp5 are required for ribosome assembly. Here, we use in vivo and in vitro biochemical analyses to demonstrate that ATP-bound Rok1, but not adenosine diphosphate (ADP)-bound Rok1, stabilizes Rrp5 binding to 40S ribosomes. Interconversion between these two forms by ATP hydrolysis is required for release of Rrp5 from pre-40S ribosomes in vivo, thereby allowing Rrp5 to carry out its role in 60S subunit assembly. Furthermore, our data also strongly suggest that the previously described accumulation of snR30 upon Rok1 inactivation arises because Rrp5 release is blocked and implicate a previously undescribed interaction between Rrp5 and the DEAD-box protein Has1 in mediating snR30 accumulation when Rrp5 release from pre-40S subunits is blocked. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6655.map.gz emd_6655.map.gz | 5.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6655-v30.xml emd-6655-v30.xml emd-6655.xml emd-6655.xml | 11.2 KB 11.2 KB | Display Display |  EMDB header EMDB header |
| Images |  400_6655.gif 400_6655.gif 80_6655.gif 80_6655.gif | 22.3 KB 6.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6655 http://ftp.pdbj.org/pub/emdb/structures/EMD-6655 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6655 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6655 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_6655.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6655.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of Rrp5 bound to RNA | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.8 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Saccharomyces cerevisiae Rrp5 bound to H45-3ITS1 RNA
| Entire | Name: Saccharomyces cerevisiae Rrp5 bound to H45-3ITS1 RNA |
|---|---|
| Components |
|
-Supramolecule #1000: Saccharomyces cerevisiae Rrp5 bound to H45-3ITS1 RNA
| Supramolecule | Name: Saccharomyces cerevisiae Rrp5 bound to H45-3ITS1 RNA / type: sample / ID: 1000 Details: At a 5-sigma contour, the mass is about 0.25 MDa, which corresponds to about 0.2 MDa from Rrp5 and 0.05 MDa from RNA. Oligomeric state: 1 / Number unique components: 2 |
|---|---|
| Molecular weight | Theoretical: 350 KDa |
-Macromolecule #1: Ribosomal RNA Processing 5
| Macromolecule | Name: Ribosomal RNA Processing 5 / type: protein_or_peptide / ID: 1 Name.synonym: Rrp5, U3 small nucleolar RNA-associated protein RRP5 Number of copies: 1 / Oligomeric state: monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Saccharomyces cerevisiae (brewer's yeast) / Strain: By 4741 / synonym: Yeast / Location in cell: nucleolus Saccharomyces cerevisiae (brewer's yeast) / Strain: By 4741 / synonym: Yeast / Location in cell: nucleolus |
| Molecular weight | Experimental: 350 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) / Recombinant strain: Rosetta2 (DE3) / Recombinant plasmid: pGEX-6-P3 Escherichia coli (E. coli) / Recombinant strain: Rosetta2 (DE3) / Recombinant plasmid: pGEX-6-P3 |
| Sequence | UniProtKB: rRNA biogenesis protein RRP5 GO:  RNA processing, RNA processing,  RNA binding, RNA binding,  protein binding, intracellular anatomical structure protein binding, intracellular anatomical structureInterPro: Nucleic acid-binding, OB-fold,  S1 domain, Tetratricopeptide-like helical domain superfamily, HAT (Half-A-TPR) repeat S1 domain, Tetratricopeptide-like helical domain superfamily, HAT (Half-A-TPR) repeat |
-Macromolecule #2: pre-rRNA
| Macromolecule | Name: pre-rRNA / type: rna / ID: 2 / Name.synonym: H45-3ITS1 RNA / Details: in vitro transcribed / Classification: OTHER / Structure: SINGLE STRANDED / Synthetic?: No |
|---|---|
| Source (natural) | Organism:   Saccharomyces cerevisiae (brewer's yeast) / Strain: By 4741 / synonym: yeast Saccharomyces cerevisiae (brewer's yeast) / Strain: By 4741 / synonym: yeast |
| Molecular weight | Theoretical: 120 KDa |
-Experimental details
-Structure determination
| Method |  negative staining negative staining |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6.7 / Details: 300 mM NaCl, 10 mM MgCl2, 30 mM MES |
|---|---|
| Staining | Type: NEGATIVE / Details: uranyl formate on floated continuous carbon |
| Grid | Details: 400 mesh copper grid, glow-discharged in Gatan Solarus apparatus |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 120 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 59000 Bright-field microscopy / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 59000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Date | Mar 23, 2013 |
| Image recording | Category: CCD / Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Number real images: 1200 / Average electron dose: 20 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final two d classification | Number classes: 50 |
|---|---|
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 30.0 Å / Resolution method: OTHER / Software - Name: EMAN Details: Final maps were calculated from two averaged datasets. Number images used: 8500 |
| Details | Particles were initially manually picked to determine de novo class averages and then automatically picked. Initial model was determined with random conical tilt. |
 Movie
Movie Controller
Controller