[English] 日本語
 Yorodumi
Yorodumi- EMDB-5698: Structure of NtrC1 ATPase in complex with Sigma-54 and promoter DNA -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5698 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
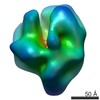
| Title | Structure of NtrC1 ATPase in complex with Sigma-54 and promoter DNA | |||||||||
 Map data Map data | Negative stained reconstruction of NtrC1 AAA+ ATPase, Sigma-54, and promoter DNA complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | AAA+ ATPase / NtrC1 /  Sigma-54 Sigma-54 | |||||||||
| Function / homology |  Function and homology information Function and homology informationDNA-binding transcription activator activity /  sigma factor activity / ribonucleoside triphosphate phosphatase activity / DNA-templated transcription initiation / DNA-directed 5'-3' RNA polymerase activity / regulation of DNA-templated transcription / sigma factor activity / ribonucleoside triphosphate phosphatase activity / DNA-templated transcription initiation / DNA-directed 5'-3' RNA polymerase activity / regulation of DNA-templated transcription /  DNA binding DNA bindingSimilarity search - Function | |||||||||
| Biological species |    Aquifex aeolicus (bacteria) / Aquifex aeolicus (bacteria) /   Klebsiella pneumoniae (bacteria) / Klebsiella pneumoniae (bacteria) /   Sinorhizobium meliloti (bacteria) Sinorhizobium meliloti (bacteria) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  negative staining / Resolution: 24.0 Å negative staining / Resolution: 24.0 Å | |||||||||
 Authors Authors | Chowdhury S / Sysoeva TA / Guo L / Nixon BT | |||||||||
 Citation Citation |  Journal: Genes Dev / Year: 2013 Journal: Genes Dev / Year: 2013Title: Nucleotide-induced asymmetry within ATPase activator ring drives σ54-RNAP interaction and ATP hydrolysis. Authors: Tatyana A Sysoeva / Saikat Chowdhury / Liang Guo / B Tracy Nixon /  Abstract: It is largely unknown how the typical homomeric ring geometry of ATPases associated with various cellular activities enables them to perform mechanical work. Small-angle solution X-ray scattering, ...It is largely unknown how the typical homomeric ring geometry of ATPases associated with various cellular activities enables them to perform mechanical work. Small-angle solution X-ray scattering, crystallography, and electron microscopy (EM) reconstructions revealed that partial ATP occupancy caused the heptameric closed ring of the bacterial enhancer-binding protein (bEBP) NtrC1 to rearrange into a hexameric split ring of striking asymmetry. The highly conserved and functionally crucial GAFTGA loops responsible for interacting with σ54-RNA polymerase formed a spiral staircase. We propose that splitting of the ensemble directs ATP hydrolysis within the oligomer, and the ring's asymmetry guides interaction between ATPase and the complex of σ54 and promoter DNA. Similarity between the structure of the transcriptional activator NtrC1 and those of distantly related helicases Rho and E1 reveals a general mechanism in homomeric ATPases whereby complex allostery within the ring geometry forms asymmetric functional states that allow these biological motors to exert directional forces on their target macromolecules. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5698.map.gz emd_5698.map.gz | 66.4 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5698-v30.xml emd-5698-v30.xml emd-5698.xml emd-5698.xml | 13 KB 13 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5698_1.tif emd_5698_1.tif | 213.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5698 http://ftp.pdbj.org/pub/emdb/structures/EMD-5698 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5698 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5698 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_5698.map.gz / Format: CCP4 / Size: 373 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5698.map.gz / Format: CCP4 / Size: 373 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Negative stained reconstruction of NtrC1 AAA+ ATPase, Sigma-54, and promoter DNA complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 6.016 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Complex of NtrC1 AAA+ ATPase hexamer with Sigma-54 and promoter DNA
| Entire | Name: Complex of NtrC1 AAA+ ATPase hexamer with Sigma-54 and promoter DNA |
|---|---|
| Components |
|
-Supramolecule #1000: Complex of NtrC1 AAA+ ATPase hexamer with Sigma-54 and promoter DNA
| Supramolecule | Name: Complex of NtrC1 AAA+ ATPase hexamer with Sigma-54 and promoter DNA type: sample / ID: 1000 / Details: The sample was monodisperse. Oligomeric state: One hexamer of NtrC1 AAA+ ATPase binds to one double-stranded promoter DNA and one Sigma-54. Number unique components: 3 |
|---|---|
| Molecular weight | Theoretical: 260 KDa |
-Macromolecule #1: ATPase Associated with various cellular activities
| Macromolecule | Name: ATPase Associated with various cellular activities / type: protein_or_peptide / ID: 1 / Name.synonym: AAA+ ATPase, Sigma-54 transcription activator / Details: Hexamer of the NtrC1 AAA+ ATPase domain / Number of copies: 6 / Oligomeric state: Hexamer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:    Aquifex aeolicus (bacteria) / Strain: VF5 Aquifex aeolicus (bacteria) / Strain: VF5 |
| Molecular weight | Theoretical: 31 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) / Recombinant strain: BL21(DE3) / Recombinant plasmid: pET122 Escherichia coli (E. coli) / Recombinant strain: BL21(DE3) / Recombinant plasmid: pET122 |
| Sequence | GO: ribonucleoside triphosphate phosphatase activity / InterPro: AAA+ ATPase domain |
-Macromolecule #2: Sigma-54
| Macromolecule | Name: Sigma-54 / type: protein_or_peptide / ID: 2 Name.synonym: Sigma-54 component of bacterial RNA polymerase Details: Single copy of bacterial transcription initiation factor Sigma-54 Number of copies: 1 / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Klebsiella pneumoniae (bacteria) Klebsiella pneumoniae (bacteria) |
| Molecular weight | Theoretical: 57 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) / Recombinant strain: BL21(DE3) / Recombinant plasmid: pET122 Escherichia coli (E. coli) / Recombinant strain: BL21(DE3) / Recombinant plasmid: pET122 |
| Sequence | UniProtKB: RNA polymerase sigma-54 factor GO: DNA-templated transcription initiation, regulation of DNA-templated transcription,  DNA binding, DNA-directed 5'-3' RNA polymerase activity, DNA binding, DNA-directed 5'-3' RNA polymerase activity,  sigma factor activity sigma factor activityInterPro: RNA polymerase sigma factor 54 |
-Macromolecule #3: nifH promoter DNA
| Macromolecule | Name: nifH promoter DNA / type: dna / ID: 3 / Name.synonym: Sigma-54 promoter DNA Details: This is a pre-melt nifH promoter, with a -11, -12 mismatch. The complementary strand has G and T at 12 and 13 base positions from 5' end instead of T and G. Classification: DNA / Structure: DOUBLE HELIX / Synthetic?: Yes |
|---|---|
| Source (natural) | Organism:   Sinorhizobium meliloti (bacteria) Sinorhizobium meliloti (bacteria) |
| Molecular weight | Theoretical: 22 KDa |
| Sequence | String: CAGACGGCTG GCACGACTTT TGCCAGATCA GCCCTG |
-Experimental details
-Structure determination
| Method |  negative staining negative staining |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.01 mg/mL |
|---|---|
| Buffer | pH: 7.9 Details: 1mM ADP, 1mM AlCl3, 8mM NaF, 1mM MgCl2, 20mM Tris-HCl, 1% (w/v) trehalose, 1mM TCEP |
| Staining | Type: NEGATIVE Details: Sample was adsorbed on thin continuous carbon coated grids, stained with 0.75% (w/v) uranyl formate, and air-dried. |
| Grid | Details: Thin carbon film on 300 mesh Cu-Rh maxtaform grids, plasma cleaned in oxygen-hydrogen gas mixture for 15s |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 2100F |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 80000 / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 50000 Bright-field microscopy / Cs: 2 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder: Room temperature specimen / Specimen holder model: JEOL / Tilt angle min: 45 / Tilt angle max: 65 |
| Date | Aug 10, 2011 |
| Image recording | Category: CCD / Film or detector model: GENERIC TVIPS (2k x 2k) / Number real images: 280 / Average electron dose: 20 e/Å2 / Camera length: 49 Details: 102 regular, untilted micrographs were collected and 89 tilt-untilt pairs of RCT micrographs were collected. |
- Image processing
Image processing
| CTF correction | Details: Per micrograph |
|---|---|
| Final angle assignment | Details: EMAN2: az 90 degrees, alt 90 degrees, phi 90 degrees |
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 24.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: XMIPP, EMAN2, SPARX Details: Final map was calculated by refinement of RCT model by iterative projection matching and back projection. Number images used: 20000 |
| Details | Particles were picked manually using XMIPP. |
 Movie
Movie Controller
Controller