[English] 日本語
 Yorodumi
Yorodumi- EMDB-5335: Sub-nanometer resolution structure of the intact T. thermophilus ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5335 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Sub-nanometer resolution structure of the intact T. thermophilus proton-driven ATP synthase reveals the arrangement of its trans-membrane helices | |||||||||
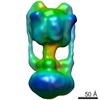
 Map data Map data | Experimental map of the V-type ATPase from Thermus thermophilus. Masks included are (a) density from subunit D not represented by the available crystal structure, (b) experimental map segment from subunit I, and (c) experimental map segment from L12-ring. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  ATP synthase / ATP synthase /  membrane protein complex / subnanometer / membrane protein complex / subnanometer /  Thermus thermophilus / Thermus thermophilus /  ATPase / ATPase /  V-ATPase V-ATPase | |||||||||
| Function / homology |  Function and homology information Function and homology informationproton-transporting V-type ATPase, V0 domain / proton-transporting two-sector ATPase complex, catalytic domain / proton-transporting ATP synthase complex / proton motive force-driven plasma membrane ATP synthesis /  H+-transporting two-sector ATPase / proton-transporting ATPase activity, rotational mechanism / proton-transporting ATP synthase activity, rotational mechanism / H+-transporting two-sector ATPase / proton-transporting ATPase activity, rotational mechanism / proton-transporting ATP synthase activity, rotational mechanism /  ATP binding / ATP binding /  metal ion binding metal ion bindingSimilarity search - Function | |||||||||
| Biological species |    Thermus thermophilus HB8 (bacteria) Thermus thermophilus HB8 (bacteria) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 9.7 Å cryo EM / Resolution: 9.7 Å | |||||||||
 Authors Authors | Lau WCY / Rubinstein JL | |||||||||
 Citation Citation |  Journal: Nature / Year: 2011 Journal: Nature / Year: 2011Title: Subnanometre-resolution structure of the intact Thermus thermophilus H+-driven ATP synthase. Authors: Wilson C Y Lau / John L Rubinstein /  Abstract: Ion-translocating rotary ATPases serve either as ATP synthases, using energy from a transmembrane ion motive force to create the cell's supply of ATP, or as transmembrane ion pumps that are powered ...Ion-translocating rotary ATPases serve either as ATP synthases, using energy from a transmembrane ion motive force to create the cell's supply of ATP, or as transmembrane ion pumps that are powered by ATP hydrolysis. The members of this family of enzymes each contain two rotary motors: one that couples ion translocation to rotation and one that couples rotation to ATP synthesis or hydrolysis. During ATP synthesis, ion translocation through the membrane-bound region of the complex causes rotation of a central rotor that drives conformational changes and ATP synthesis in the catalytic region of the complex. There are no structural models available for the intact membrane region of any ion-translocating rotary ATPase. Here we present a 9.7 Å resolution map of the H(+)-driven ATP synthase from Thermus thermophilus obtained by electron cryomicroscopy of single particles in ice. The 600-kilodalton complex has an overall subunit composition of A(3)B(3)CDE(2)FG(2)IL(12). The membrane-bound motor consists of a ring of L subunits and the carboxy-terminal region of subunit I, which are equivalent to the c and a subunits of most other rotary ATPases, respectively. The map shows that the ring contains 12 L subunits and that the I subunit has eight transmembrane helices. The L(12) ring and I subunit have a surprisingly small contact area in the middle of the membrane, with helices from the I subunit making contacts with two different L subunits. The transmembrane helices of subunit I form bundles that could serve as half-channels across the membrane, with the first half-channel conducting protons from the periplasm to the L(12) ring and the second half-channel conducting protons from the L(12) ring to the cytoplasm. This structure therefore suggests the mechanism by which a transmembrane proton motive force is converted to rotation in rotary ATPases. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5335.map.gz emd_5335.map.gz | 59.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5335-v30.xml emd-5335-v30.xml emd-5335.xml emd-5335.xml | 20.3 KB 20.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5335_1.png emd_5335_1.png | 120.5 KB | ||
| Masks |  emd_5335_msk_1.map emd_5335_msk_1.map emd_5335_msk_2.map emd_5335_msk_2.map emd_5335_msk_3.map emd_5335_msk_3.map | 64 MB 64 MB 64 MB |  Mask map Mask map | |
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5335 http://ftp.pdbj.org/pub/emdb/structures/EMD-5335 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5335 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5335 | HTTPS FTP |
-Related structure data
| Related structure data |  3j0jMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_5335.map.gz / Format: CCP4 / Size: 62.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5335.map.gz / Format: CCP4 / Size: 62.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Experimental map of the V-type ATPase from Thermus thermophilus. Masks included are (a) density from subunit D not represented by the available crystal structure, (b) experimental map segment from subunit I, and (c) experimental map segment from L12-ring. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.4 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Segmentation: Experimental map segment from subunit I
| Annotation | Experimental map segment from subunit I | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| File |  emd_5335_msk_1.map emd_5335_msk_1.map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Segmentation: Experimental map segment from L12-ring
| Annotation | Experimental map segment from L12-ring | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| File |  emd_5335_msk_2.map emd_5335_msk_2.map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Segmentation: Density from subunit D not represented by the...
| Annotation | Density from subunit D not represented by the available crystal structure | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| File |  emd_5335_msk_3.map emd_5335_msk_3.map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Thermus thermophilus V-type ATPase/ATP synthase solubilized with ...
| Entire | Name: Thermus thermophilus V-type ATPase/ATP synthase solubilized with detergent |
|---|---|
| Components |
|
-Supramolecule #1000: Thermus thermophilus V-type ATPase/ATP synthase solubilized with ...
| Supramolecule | Name: Thermus thermophilus V-type ATPase/ATP synthase solubilized with detergent type: sample / ID: 1000 / Details: Solubilized with the detergent dodecyl maltoside / Oligomeric state: Monomeric / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 600 KDa / Theoretical: 600 KDa |
-Macromolecule #1: V-type ATPase
| Macromolecule | Name: V-type ATPase / type: protein_or_peptide / ID: 1 / Name.synonym: V-type ATPase / Number of copies: 1 / Oligomeric state: monomer / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:    Thermus thermophilus HB8 (bacteria) / synonym: Thermus thermophilus / Cell: Thermus thermophilus HB8 / Organelle: plasma membrane / Location in cell: plasma membrane Thermus thermophilus HB8 (bacteria) / synonym: Thermus thermophilus / Cell: Thermus thermophilus HB8 / Organelle: plasma membrane / Location in cell: plasma membrane |
| Molecular weight | Theoretical: 600 KDa |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.5 mg/mL |
|---|---|
| Buffer | pH: 8 Details: 50mM Tris-HCl, 5mM MgCl2, 150 mM NaCl, 0.02% (w/v) dodecyl maltoside |
| Grid | Details: 200 mesh quantifoil |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 95 K / Instrument: FEI VITROBOT MARK III / Details: Vitrification instrument: Vitrobot Mark 3 / Method: Blot for 20 seconds before plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 50000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.0 mm / Nominal defocus max: 4.5 µm / Nominal defocus min: 2.5 µm / Nominal magnification: 50000 Bright-field microscopy / Cs: 2.0 mm / Nominal defocus max: 4.5 µm / Nominal defocus min: 2.5 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder: Gatan 626 / Specimen holder model: GATAN LIQUID NITROGEN |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism corrected around 100,000x magnification |
| Date | Nov 3, 2001 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 7 µm / Number real images: 700 / Average electron dose: 18 e/Å2 / Bits/pixel: 8 |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Details: Each particle |
|---|---|
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 9.7 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: Frealign, Refine_Fspace, Build_FSpace Details: Map constructed in Fourier space by Build_Fspace, sharpened with an inverse temperature factor of 750 Angstroms squared Number images used: 46105 |
-Atomic model buiding 1
| Initial model | PDB ID: Chain - #0 - Chain ID: I / Chain - #1 - Chain ID: J / Chain - #2 - Chain ID: K / Chain - #3 - Chain ID: L / Chain - #4 - Chain ID: M / Chain - #5 - Chain ID: N / Chain - #6 - Chain ID: O / Chain - #7 - Chain ID: P |
|---|---|
| Software | Name:  UCSF Chimera UCSF Chimera |
| Details | PDBEntryID_givenInChain. Protocol: Rigid Body. V1 subcomplex crystal structure |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: Correlation coefficient |
| Output model |  PDB-3j0j: |
-Atomic model buiding 2
| Initial model | PDB ID: Chain - Chain ID: A |
|---|---|
| Software | Name: Imodfit |
| Details | PDBEntryID_givenInChain. Protocol: Flexible. C subunit crystal structure |
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
| Output model |  PDB-3j0j: |
-Atomic model buiding 3
| Initial model | PDB ID: Chain - #0 - Chain ID: G / Chain - #1 - Chain ID: E |
|---|---|
| Software | Name:  UCSF Chimera UCSF Chimera |
| Details | PDBEntryID_givenInChain. Protocol: Rigid Body. Two copies of this peripheral stalk (EG subcomplex) structure fitted into the cryo-EM map because there are two copies in the complex. |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: Correlation coefficient |
| Output model |  PDB-3j0j: |
 Movie
Movie Controller
Controller













 Z
Z Y
Y X
X




























