[English] 日本語
 Yorodumi
Yorodumi- EMDB-5278: 3D structure of a full-length type 1 inositol 1,4,5-trisphosphate... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5278 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
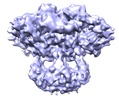
| Title | 3D structure of a full-length type 1 inositol 1,4,5-trisphosphate receptor in the closed state | |||||||||
 Map data Map data | This is a Cryo-EM density map of IP3R1 in the closed state. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | type 1 inositol 1 / 4 / 5-trisphosphate receptor /  calcium channel / closed state / single particle cryo-EM calcium channel / closed state / single particle cryo-EM | |||||||||
| Function / homology | RIH domain / endoplasmic reticulum membrane Function and homology information Function and homology information | |||||||||
| Biological species |   Rattus norvegicus (Norway rat) Rattus norvegicus (Norway rat) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / cryo EM /  negative staining / Resolution: 9.5 Å negative staining / Resolution: 9.5 Å | |||||||||
 Authors Authors | Ludtke SJ / Tran TP / Ngo QT / Moiseenkova-Bell VY / Chiu W / Serysheva II | |||||||||
 Citation Citation |  Journal: Structure / Year: 2011 Journal: Structure / Year: 2011Title: Flexible architecture of IP3R1 by Cryo-EM. Authors: Steven J Ludtke / Thao P Tran / Que T Ngo / Vera Yu Moiseenkova-Bell / Wah Chiu / Irina I Serysheva /  Abstract: Inositol 1,4,5-trisphosphate receptors (IP3Rs) play a fundamental role in generating Ca2+ signals that trigger many cellular processes in virtually all eukaryotic cells. Thus far, the three- ...Inositol 1,4,5-trisphosphate receptors (IP3Rs) play a fundamental role in generating Ca2+ signals that trigger many cellular processes in virtually all eukaryotic cells. Thus far, the three-dimensional (3D) structure of these channels has remained extremely controversial. Here, we report a subnanometer resolution electron cryomicroscopy (cryo-EM) structure of a fully functional type 1 IP3R from cerebellum in the closed state. The transmembrane region reveals a twisted bundle of four α helices, one from each subunit, that form a funnel shaped structure around the 4-fold symmetry axis, strikingly similar to the ion-conduction pore of K+ channels. The lumenal face of IP3R1 has prominent densities that surround the pore entrance and similar to the highly structured turrets of Kir channels. 3D statistical analysis of the cryo-EM density map identifies high variance in the cytoplasmic region. This structural variation could be attributed to genuine structural flexibility of IP3R1. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5278.map.gz emd_5278.map.gz | 56.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5278-v30.xml emd-5278-v30.xml emd-5278.xml emd-5278.xml | 10.7 KB 10.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5278.png emd_5278.png | 1.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5278 http://ftp.pdbj.org/pub/emdb/structures/EMD-5278 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5278 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5278 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_5278.map.gz / Format: CCP4 / Size: 62.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5278.map.gz / Format: CCP4 / Size: 62.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is a Cryo-EM density map of IP3R1 in the closed state. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.81 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Type 1 Inositol 1,4,5-Trisphosphate Receptor
| Entire | Name: Type 1 Inositol 1,4,5-Trisphosphate Receptor |
|---|---|
| Components |
|
-Supramolecule #1000: Type 1 Inositol 1,4,5-Trisphosphate Receptor
| Supramolecule | Name: Type 1 Inositol 1,4,5-Trisphosphate Receptor / type: sample / ID: 1000 Details: The sample was frozen immediately upon channel protein purification Oligomeric state: Tetramer / Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 1.3 MDa |
-Macromolecule #1: Ion channel
| Macromolecule | Name: Ion channel / type: protein_or_peptide / ID: 1 / Name.synonym: IP3R1 Details: IP3R1 channel was solubilized with CHAPS from rat cerebellum and purified by immunoaffinity chromatography. Number of copies: 4 / Oligomeric state: Tetramer / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:   Rattus norvegicus (Norway rat) / synonym: Rat / Tissue: Cerebellum / Organelle: Endoplasmic Reticulum Membranes / Location in cell: Integral membrane protein Rattus norvegicus (Norway rat) / synonym: Rat / Tissue: Cerebellum / Organelle: Endoplasmic Reticulum Membranes / Location in cell: Integral membrane protein |
| Molecular weight | Theoretical: 1.3 MDa |
| Sequence | GO: endoplasmic reticulum membrane / InterPro: RIH domain |
-Experimental details
-Structure determination
| Method |  negative staining, negative staining,  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 7.4 Details: 50 mM Tris-HCl, 150 mM sodium chloride, 1 mM DTT, 1 mM EDTA, 0.4% CHAPS, 5% sucrose, protease inhibitors |
| Staining | Type: NEGATIVE / Details: Frozen-hydrated |
| Grid | Details: 400 mesh Quantifoil holey grids covered with a continuous carbon film |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 101 K / Instrument: OTHER / Details: Vitrification instrument: Vitrobot (FEI) / Method: Blot for 2-3 sec before plunging |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 2010F |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.0 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.4 µm / Nominal magnification: 60000 Bright-field microscopy / Cs: 2.0 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.4 µm / Nominal magnification: 60000 |
| Sample stage | Specimen holder: Side entry liquid nitrogen-cooled / Specimen holder model: GATAN LIQUID NITROGEN |
| Temperature | Min: 103 K / Max: 104 K / Average: 103 K |
| Image recording | Category: CCD / Film or detector model: GENERIC GATAN (4k x 4k) / Digitization - Sampling interval: 1.81 µm / Number real images: 869 / Average electron dose: 18 e/Å2 / Bits/pixel: 16 |
- Image processing
Image processing
| CTF correction | Details: Per particle phase-flipping with amplitude correction of class-averages |
|---|---|
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 9.5 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: EMAN1.9 Details: Direct Fourier Inversion based on Wiener-filtered and CTF amplitude-corrected class-averages Number images used: 37231 |
| Details | 869 CCD frames |
 Movie
Movie Controller
Controller