[English] 日本語
 Yorodumi
Yorodumi- EMDB-5009: A cryo-EM map of the FimD-tip complex, a bacterial surface pilus ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5009 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
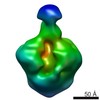
| Title | A cryo-EM map of the FimD-tip complex, a bacterial surface pilus assembly intermediate in complex with the outer membrane secretion channel. | |||||||||
 Map data Map data | 3D Cryo-EM map of FimD-tip complex, a bacterial outer membrane pilus assembly intermediate | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  cryo-electron microscopy / bacterial pilus / bacterial outer membrane secretion channel / pilus biogenesis cryo-electron microscopy / bacterial pilus / bacterial outer membrane secretion channel / pilus biogenesis | |||||||||
| Biological species |   Escherichia coli (E. coli) Escherichia coli (E. coli) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 23.0 Å cryo EM / Resolution: 23.0 Å | |||||||||
 Authors Authors | Tang C / Thanassi D / Li H | |||||||||
 Citation Citation |  Journal: Cell / Year: 2008 Journal: Cell / Year: 2008Title: Fiber formation across the bacterial outer membrane by the chaperone/usher pathway. Authors: Han Remaut / Chunyan Tang / Nadine S Henderson / Jerome S Pinkner / Tao Wang / Scott J Hultgren / David G Thanassi / Gabriel Waksman / Huilin Li /  Abstract: Gram-negative pathogens commonly exhibit adhesive pili on their surfaces that mediate specific attachment to the host. A major class of pili is assembled via the chaperone/usher pathway. Here, the ...Gram-negative pathogens commonly exhibit adhesive pili on their surfaces that mediate specific attachment to the host. A major class of pili is assembled via the chaperone/usher pathway. Here, the structural basis for pilus fiber assembly and secretion performed by the outer membrane assembly platform--the usher--is revealed by the crystal structure of the translocation domain of the P pilus usher PapC and single particle cryo-electron microscopy imaging of the FimD usher bound to a translocating type 1 pilus assembly intermediate. These structures provide molecular snapshots of a twinned-pore translocation machinery in action. Unexpectedly, only one pore is used for secretion, while both usher protomers are used for chaperone-subunit complex recruitment. The translocating pore itself comprises 24 beta strands and is occluded by a folded plug domain, likely gated by a conformationally constrained beta-hairpin. These structures capture the secretion of a virulence factor across the outer membrane of gram-negative bacteria. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5009.map.gz emd_5009.map.gz | 3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5009-v30.xml emd-5009-v30.xml emd-5009.xml emd-5009.xml | 11.8 KB 11.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5009_1.tif emd_5009_1.tif | 750.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5009 http://ftp.pdbj.org/pub/emdb/structures/EMD-5009 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5009 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5009 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_5009.map.gz / Format: CCP4 / Size: 3.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5009.map.gz / Format: CCP4 / Size: 3.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D Cryo-EM map of FimD-tip complex, a bacterial outer membrane pilus assembly intermediate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.54 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : FimD-tip complex
| Entire | Name: FimD-tip complex |
|---|---|
| Components |
|
-Supramolecule #1000: FimD-tip complex
| Supramolecule | Name: FimD-tip complex / type: sample / ID: 1000 / Details: The sample was monodisperse Oligomeric state: FimD usher dimer in complex with one copy each of FimH, FimF, FimG pilins and FimC chaperone Number unique components: 5 |
|---|---|
| Molecular weight | Experimental: 260 KDa / Theoretical: 260 KDa |
-Supramolecule #1: FimD-tip complex
| Supramolecule | Name: FimD-tip complex / type: organelle_or_cellular_component / ID: 1 / Name.synonym: Pilus assembly usher / Details: This component forms a dimer in the complex / Number of copies: 1 / Oligomeric state: Dimer / Recombinant expression: Yes |
|---|---|
| Ref GO | divclassse qspanoncli ckpopupspa nclassgree n(this)spandata popltspanc lassquotlo adingbarqu otgtltimgs rcquotimgl oadinggifq uotdecodin gquotasync quotgtltsp angtdataur lajaxphp?m odetaxoamp ... divclassse qspanoncli ckpopupspa nclassgree n(this)spandata popltspanc lassquotlo adingbarqu otgtltimgs rcquotimgl oadinggifq uotdecodin gquotasync quotgtltsp angtdataur lajaxphp?m odetaxoamp kGO3A00154 73ampajax1 classpoptr giGO001547 3ispandiv |
| Ref INTERPRO | divclassse qspanoncli ckpopupspa nclassgree n(this)spandata popltspanc lassquotlo adingbarqu otgtltimgs rcquotimgl oadinggifq uotdecodin gquotasync quotgtltsp angtdataur lajaxphp?m odetaxoamp ... divclassse qspanoncli ckpopupspa nclassgree n(this)spandata popltspanc lassquotlo adingbarqu otgtltimgs rcquotimgl oadinggifq uotdecodin gquotasync quotgtltsp angtdataur lajaxphp?m odetaxoamp kIPR000015 ampajax1cl asspoptrgi IPR000015i spandiv |
| Source (natural) | Organism:   Escherichia coli (E. coli) / Location in cell: Outer membrane Escherichia coli (E. coli) / Location in cell: Outer membrane |
| Molecular weight | Experimental: 96 KDa / Theoretical: 96 KDa |
| Recombinant expression | Organism:  Escherichia coli B strain Tuner (Novagen) / Recombinant plasmid: Tuner/pAN2 and pNH237 Escherichia coli B strain Tuner (Novagen) / Recombinant plasmid: Tuner/pAN2 and pNH237 |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.04 mg/mL |
|---|---|
| Buffer | pH: 8 / Details: 20 mM Tris-HCl (pH 8), 0.15 M NaCl, 0.05% DDM. |
| Grid | Details: glow-discharged lacey carbon grid |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 85 % / Chamber temperature: 108 K / Instrument: OTHER Details: Vitrification instrument: Vitrobot. 12 degree Celsius chamber temperature Method: 6 seconds blot |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 2010F |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.0 mm / Nominal defocus max: 5.0 µm / Nominal defocus min: 3.0 µm / Nominal magnification: 50000 Bright-field microscopy / Cs: 2.0 mm / Nominal defocus max: 5.0 µm / Nominal defocus min: 3.0 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder: Gatan 626 cryo holder / Specimen holder model: GATAN LIQUID NITROGEN |
| Temperature | Min: 100 K / Max: 105 K / Average: 103 K |
| Alignment procedure | Legacy - Astigmatism: correction at 250,000 mag / Legacy - Electron beam tilt params: -2 mrad |
| Date | Feb 1, 2007 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: NIKON SUPER COOLSCAN 9000 / Digitization - Sampling interval: 12.7 µm / Number real images: 100 / Average electron dose: 10 e/Å2 / Od range: 1.3 / Bits/pixel: 14 |
- Image processing
Image processing
| CTF correction | Details: Each films |
|---|---|
| Final two d classification | Number classes: 100 |
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 23.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: EMAN, SPIDER / Number images used: 11000 |
| Details | particles were manually selected |
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | PDBEntryID_givenInChain. Protocol: Rigid Body. manual docking followed by local correlation based real space fitting in chimera |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: correlation |
 Movie
Movie Controller
Controller