[English] 日本語
 Yorodumi
Yorodumi- EMDB-43893: Structure of the auto-fluorescent membrane-bound red body organel... -
+ Open data
Open data
- Basic information
Basic information
| Entry | 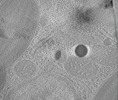 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the auto-fluorescent membrane-bound red body organelle from Nannochloropsis oceanica in situ | |||||||||
 Map data Map data | Tomographic reconstruction of a cryo-lamella through a Nannochloropsis oceanica cell showing a red body within its cellular context. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  Organelle / Algae / Organelle / Algae /  Intracellular Transport / Intracellular Transport /  Carotenoid / LIPID TRANSPORT Carotenoid / LIPID TRANSPORT | |||||||||
| Biological species |  Nannochloropsis oceanica (eukaryote) Nannochloropsis oceanica (eukaryote) | |||||||||
| Method |  electron tomography / electron tomography /  cryo EM cryo EM | |||||||||
 Authors Authors | Grob P / Danielle J / Gee CW | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: A proposed function for the red body of Nannochloropsis in the formation of the recalcitrant cell wall polymer, algaenan Authors: Gee CW / Andersen-Ranberg J | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_43893.map.gz emd_43893.map.gz | 514.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-43893-v30.xml emd-43893-v30.xml emd-43893.xml emd-43893.xml | 11.8 KB 11.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_43893.png emd_43893.png | 199.2 KB | ||
| Filedesc metadata |  emd-43893.cif.gz emd-43893.cif.gz | 4.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43893 http://ftp.pdbj.org/pub/emdb/structures/EMD-43893 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43893 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43893 | HTTPS FTP |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_43893.map.gz / Format: CCP4 / Size: 558.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_43893.map.gz / Format: CCP4 / Size: 558.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Tomographic reconstruction of a cryo-lamella through a Nannochloropsis oceanica cell showing a red body within its cellular context. | ||||||||||||||||||||
| Voxel size | X=Y=Z: 22.13 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Nannochloropsis oceanica CCMP1779
| Entire | Name: Nannochloropsis oceanica CCMP1779 |
|---|---|
| Components |
|
-Supramolecule #1: Nannochloropsis oceanica CCMP1779
| Supramolecule | Name: Nannochloropsis oceanica CCMP1779 / type: cell / ID: 1 / Parent: 0 Details: Tomographic reconstruction of a red body organelle from Nannochloropsis oceanica in situ |
|---|---|
| Source (natural) | Organism:  Nannochloropsis oceanica (eukaryote) / Strain: CCMP1779 Nannochloropsis oceanica (eukaryote) / Strain: CCMP1779 |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  electron tomography electron tomography |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 8.1 Component:
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R2/2 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Support film - Film thickness: 12 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.027 kPa / Details: the grid was soaked in chloroform before use | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV / Details: Blotted manually from opposite side of the grid. | ||||||||||||
| Details | Nannochloropsis oceanica CCMP1779 cells were grown in artificial seawater and f-media enrichment entrained to a 12-12 light-dark photoperiod and sampled in mid-log phase (~1x10^7 cells/ml), shortly after subjective dark when cells are undergoing division. | ||||||||||||
| Sectioning | Focused ion beam - Instrument: OTHER / Focused ion beam - Ion: OTHER / Focused ion beam - Voltage: 30 / Focused ion beam - Current: 0.037 / Focused ion beam - Duration: 600 / Focused ion beam - Temperature: 123 K / Focused ion beam - Initial thickness: 1000 / Focused ion beam - Final thickness: 380 Focused ion beam - Details: Grids were transfered to a Leica Ace 900 (Leica Microsystems) for coating with 5nm of platinum prior to being transferred to the Zeiss Crossbeam 540 (Zeiss, Germany). The ...Focused ion beam - Details: Grids were transfered to a Leica Ace 900 (Leica Microsystems) for coating with 5nm of platinum prior to being transferred to the Zeiss Crossbeam 540 (Zeiss, Germany). The Zeiss Crossbeam 540 operated with a Leica CryoStage (Leica Microsystems, GmbH) cooled to -150 deg. C was used for milling and imaging of the frozen cells. To create lamellae used for cryo-tomography, milling was performed with a gallium ion source at an energy of 37 pA and a working distance of 5mm. Imaging of the grid and milled lamellae was done using an Everhart-Thornley detector at 2.0 kV.. The value given for _em_focused_ion_beam.instrument is Zeiss Crossbeam 540. This is not in a list of allowed values {'DB235', 'OTHER'} so OTHER is written into the XML file. |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 6.0 µm / Nominal defocus min: 1.6 µm / Nominal magnification: 15000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 6.0 µm / Nominal defocus min: 1.6 µm / Nominal magnification: 15000 |
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 35 eV |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 100.0 e/Å2 Details: images were collected in movie mode and super resolution |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Algorithm: BACK PROJECTION / Software - Name:  IMOD (ver. 4.11.24) / Software - details: Etomo / Number images used: 61 IMOD (ver. 4.11.24) / Software - details: Etomo / Number images used: 61 |
|---|---|
| Details | the movie frames were aligned with MotionCor2 and binned to normal resolution |
 Movie
Movie Controller
Controller



