+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | TehA from Haemophilus influenzae purified in LMNG | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  ANION CHANNEL / ALPHA HELICAL INTEGRAL MEMBRANE PROTEIN / ANION CHANNEL / ALPHA HELICAL INTEGRAL MEMBRANE PROTEIN /  MEMBRANE PROTEIN MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationmonoatomic cation efflux transmembrane transporter activity / response to tellurium ion / response to antibiotic / identical protein binding /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |   Haemophilus influenzae (bacteria) Haemophilus influenzae (bacteria) | |||||||||
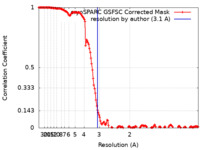
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.1 Å cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Catalano C / Senko S / Tran NL / Lucier KW / Farwell AC / Silva MS / Dip PV / Poweleit N / Scapin G | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Int J Mol Sci / Year: 2024 Journal: Int J Mol Sci / Year: 2024Title: High-Resolution Cryo-Electron Microscopy Structure Determination of Tellurite-Resistance Protein A via 200 kV Transmission Electron Microscopy. Authors: Nhi L Tran / Skerdi Senko / Kyle W Lucier / Ashlyn C Farwell / Sabrina M Silva / Phat V Dip / Nicole Poweleit / Giovanna Scapin / Claudio Catalano /  Abstract: Membrane proteins constitute about 20% of the human proteome and play crucial roles in cellular functions. However, a complete understanding of their structure and function is limited by their ...Membrane proteins constitute about 20% of the human proteome and play crucial roles in cellular functions. However, a complete understanding of their structure and function is limited by their hydrophobic nature, which poses significant challenges in purification and stabilization. Detergents, essential in the isolation process, risk destabilizing or altering the proteins' native conformations, thus affecting stability and functionality. This study leverages single-particle cryo-electron microscopy to elucidate the structural nuances of membrane proteins, focusing on the SLAC1 bacterial homolog from (TehA) purified with diverse detergents, including n-dodecyl β-D-maltopyranoside (DDM), glycodiosgenin (GDN), β-D-octyl-glucoside (OG), and lauryl maltose neopentyl glycol (LMNG). This research not only contributes to the understanding of membrane protein structures but also addresses detergent effects on protein purification. By showcasing that the overall structural integrity of the channel is preserved, our study underscores the intricate interplay between proteins and detergents, offering insightful implications for drug design and membrane biology. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_43248.map.gz emd_43248.map.gz | 306.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-43248-v30.xml emd-43248-v30.xml emd-43248.xml emd-43248.xml | 16.2 KB 16.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_43248_fsc.xml emd_43248_fsc.xml | 14.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_43248.png emd_43248.png | 39.4 KB | ||
| Masks |  emd_43248_msk_1.map emd_43248_msk_1.map | 325 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-43248.cif.gz emd-43248.cif.gz | 5.7 KB | ||
| Others |  emd_43248_half_map_1.map.gz emd_43248_half_map_1.map.gz emd_43248_half_map_2.map.gz emd_43248_half_map_2.map.gz | 301.7 MB 301.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43248 http://ftp.pdbj.org/pub/emdb/structures/EMD-43248 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43248 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43248 | HTTPS FTP |
-Related structure data
| Related structure data |  8vi4MC  8vi2C  8vi3C  8vi5C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_43248.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_43248.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.566 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_43248_msk_1.map emd_43248_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_43248_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_43248_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : TELLURITE RESISTANCE PROTEIN TEHA
| Entire | Name: TELLURITE RESISTANCE PROTEIN TEHA |
|---|---|
| Components |
|
-Supramolecule #1: TELLURITE RESISTANCE PROTEIN TEHA
| Supramolecule | Name: TELLURITE RESISTANCE PROTEIN TEHA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Haemophilus influenzae (bacteria) Haemophilus influenzae (bacteria) |
| Molecular weight | Theoretical: 108 KDa |
-Macromolecule #1: Tellurite resistance protein TehA homolog
| Macromolecule | Name: Tellurite resistance protein TehA homolog / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Haemophilus influenzae (bacteria) Haemophilus influenzae (bacteria) |
| Molecular weight | Theoretical: 35.252547 KDa |
| Recombinant expression | Organism:   Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) |
| Sequence | String: MNITKPFPLP TGYFGIPLGL AALSLAWFHL ENLFPAARMV SDVLGIVASA VWILFILMYA YKLRYYFEEV RAEYHSPVRF SFIALIPIT TMLVGDILYR WNPLIAEVLI WIGTIGQLLF STLRVSELWQ GGVFEQKSTH PSFYLPAVAA NFTSASSLAL L GYHDLGYL ...String: MNITKPFPLP TGYFGIPLGL AALSLAWFHL ENLFPAARMV SDVLGIVASA VWILFILMYA YKLRYYFEEV RAEYHSPVRF SFIALIPIT TMLVGDILYR WNPLIAEVLI WIGTIGQLLF STLRVSELWQ GGVFEQKSTH PSFYLPAVAA NFTSASSLAL L GYHDLGYL FFGAGMIAWI IFEPVLLQHL RISSLEPQFR ATMGIVLAPA FVCVSAYLSI NHGEVDTLAK ILWGYGFLQL FF LLRLFPW IVEKGLNIGL WAFSFGLASM ANSATAFYHG NVLQGVSIFA FVFSNVMIGL LVLMTIYKLT KGQFFLK UniProtKB: Tellurite resistance protein TehA homolog |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 240000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 240000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number real images: 11370 / Average electron dose: 36.43 e/Å2 |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)