[English] 日本語
 Yorodumi
Yorodumi- EMDB-42949: Microtubule inner proteins in the 48-nm doublet microtubule from ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Microtubule inner proteins in the 48-nm doublet microtubule from the proximal region of Tetrahymena thermophila strain K40R | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  Cilia / Cilia /  Axoneme / Doublet Microtubule / Microtubule Inner Protein / Axoneme / Doublet Microtubule / Microtubule Inner Protein /  STRUCTURAL PROTEIN STRUCTURAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |   Tetrahymena thermophila (eukaryote) Tetrahymena thermophila (eukaryote) | |||||||||
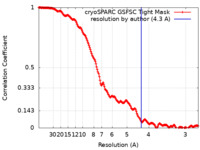
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 4.3 Å cryo EM / Resolution: 4.3 Å | |||||||||
 Authors Authors | Legal T / Bui KH / Yang SK | |||||||||
| Funding support |  Canada, 2 items Canada, 2 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: 48-nm doublet microtubule from the proximal region of Tetrahymena thermophila strain K40R Authors: Bui KH | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42949.map.gz emd_42949.map.gz | 463.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42949-v30.xml emd-42949-v30.xml emd-42949.xml emd-42949.xml | 17.1 KB 17.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_42949_fsc.xml emd_42949_fsc.xml | 16.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_42949.png emd_42949.png | 146.6 KB | ||
| Masks |  emd_42949_msk_1.map emd_42949_msk_1.map | 512 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-42949.cif.gz emd-42949.cif.gz | 6.6 KB | ||
| Others |  emd_42949_half_map_1.map.gz emd_42949_half_map_1.map.gz emd_42949_half_map_2.map.gz emd_42949_half_map_2.map.gz | 475.4 MB 475.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42949 http://ftp.pdbj.org/pub/emdb/structures/EMD-42949 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42949 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42949 | HTTPS FTP |
-Related structure data
| Related structure data |  8v3lMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_42949.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42949.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.37 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_42949_msk_1.map emd_42949_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_42949_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_42949_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : 48-nm proximal repeat unit of microtubule doublet from Tetrahymen...
| Entire | Name: 48-nm proximal repeat unit of microtubule doublet from Tetrahymena thermophila strain K40R |
|---|---|
| Components |
|
-Supramolecule #1: 48-nm proximal repeat unit of microtubule doublet from Tetrahymen...
| Supramolecule | Name: 48-nm proximal repeat unit of microtubule doublet from Tetrahymena thermophila strain K40R type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all Details: Single Particle Analysis of the proximal 48-nm repeating unit of the axoneme from intact cilia purified from Tetrahymena strain K40R |
|---|---|
| Source (natural) | Organism:   Tetrahymena thermophila (eukaryote) / Strain: K40R / Organelle: Cilia Tetrahymena thermophila (eukaryote) / Strain: K40R / Organelle: Cilia |
| Molecular weight | Theoretical: 7.6 MDa |
-Macromolecule #1: CCDC81B
| Macromolecule | Name: CCDC81B / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Tetrahymena thermophila (eukaryote) / Strain: K40R Tetrahymena thermophila (eukaryote) / Strain: K40R |
| Molecular weight | Theoretical: 179.311766 KDa |
| Sequence | String: MRSVISEQSL ASTKTQKLFN NLISKGQSKI PLLEGQLDEE SLTKTWGAFG LYLSRQLRMG RGTNIPHFGT FTFSAPECNL TGVTNPIER DKQFRVPVFL VSKEFVLGQP IKTGIFIDKH IRPYNVQTSG SISVTKINFT EIAQYADQNK EAVKISFERI I RQLADQTR ...String: MRSVISEQSL ASTKTQKLFN NLISKGQSKI PLLEGQLDEE SLTKTWGAFG LYLSRQLRMG RGTNIPHFGT FTFSAPECNL TGVTNPIER DKQFRVPVFL VSKEFVLGQP IKTGIFIDKH IRPYNVQTSG SISVTKINFT EIAQYADQNK EAVKISFERI I RQLADQTR QKRVVDMEIP GIGVFQVRNN IAAVHFDEYL IQNIKGVPKR PLSERKQKGD MNLTADRLSM LENTSRIGAD AE QYLKTKL GIDIDNISTK SRPLTAVRNA STSNLLQPNN KTLQRPATST TLRQSAINLK QPELGPLEFG LKRLKFWIRQ NCF NAQDAF LEMCNCALGR TRAQRVKVDY KQFFEAINKM ELDLNEEQIQ ELFKQLDANK DGYINYTDWR NTIKDDNDHL QYIK DVIYK RQLHTDDVLK TMGLDREHPP INKYQLKDAL KKLDLTLTDI KAIRVAQTLL GHEEYISMYD LARSFETIEE DDKIF DPSW FKDILYKIKQ KIMALENPNE IRESFENYDE HNEGILDTAN FKTCLMRSQL KLTVKEINRI VRYLPKVKVN NIDYYN FLK LVERVDINAS APDAVSDISE FAVKLGKFIK DKKFTIPSFL KTVRRFSFVL DSSNNQDYFV TLTQMSEFLH RNVFKGN DR LDIDFYVNEL DIDCDGYIKE NDVQSFLNRY TYFDQTQYTP LSMTRSLTLK SLDPRTVSMN TKTLYPFKAL SEAKVDTI L RDLRKKMELK KMKAAELFST LDADQDGFLT INEFSENIRK IIELSQPAID GFFAYMDKLH TGMVDLSSFL KVMRKSIVT KDQPIKEDNF DWENEMVFRI RQWFKKEGIT VEDSFRAIDQ NFKREINVDD LEIFLKDCLK VKSEELNKAK LDRLFKLIDQ YKRGKVNFV DWKRFIMEDF GYGMNQTIMG GKKIDNISSF DWKINARQQL GLVLTRRFST LSHSFDVISG YRKKLLFSHF K KWVQDTDA LRGFDLTESL MQELFSDLDS HNKGYLSESD WEIAFGGFDW EHQMHEEIKE YVKQNFKSIE KAFEFFCQGN QN KIIEQAD FNKAINSILP KRFIDSDLKD LWVYVSSGQP QVDFHRFKIA FQKYIPHYER TIEYDQENMR PKTAGPLSSL SKS TSWNNI YGASQLNGNS QNPFGTSSIV DGNEDDVFEE KLLSKIRRLL RPTIKSVKKV LEQLDVNNIG YITNLEFRNA IKTL NLGLA QNEVDLLLNI CEIDGRVNIK QFLKFIDFSE SDKKIVQRSI LKLREITNQV YTFLLSPKDA FRMFDVDSNG YLDFD QFSQ FIGKLSQLSG HDMLPYSIIK DLFEYIDSKK DGVIDQAEWM DIFNKFEYVP QASINKQPLS KATVYNLQKR PQTSFN KST LNELPHNKFN LEYISSNYPL TVKNEKIHQI SDKEKYGTMN KFFDEVERQP ESKPVQDNHF NQILGEWGKT KEFDKII IS IGKNRRFLLD MFGSLQQRNI PITFERAKAI IGQMLRNSGQ SLKEESWPII LKFAERNGLI DYKFLLEVYK ERIKRIDS H PIMSRNRSLQ KFTS UniProtKB:  EF-hand protein EF-hand protein |
-Macromolecule #2: BMIP1
| Macromolecule | Name: BMIP1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Tetrahymena thermophila (eukaryote) / Strain: K40R Tetrahymena thermophila (eukaryote) / Strain: K40R |
| Molecular weight | Theoretical: 28.564789 KDa |
| Sequence | String: MQSTITSTTS LTPQQILNQI IKPEYIKYYS DWIHTANEKE ARGLQLLGAI YKHKGSKKFR AKPPSVQLQQ LHAQQEFDFQ KAEEMYRKS QITSSYGQEF GYLQKDLKPT QNVFKYKKCS DLPFAKPLSD LAKLFIDNWI ELNDELEFQE LVLACLRSLH S RCIAQEVP ...String: MQSTITSTTS LTPQQILNQI IKPEYIKYYS DWIHTANEKE ARGLQLLGAI YKHKGSKKFR AKPPSVQLQQ LHAQQEFDFQ KAEEMYRKS QITSSYGQEF GYLQKDLKPT QNVFKYKKCS DLPFAKPLSD LAKLFIDNWI ELNDELEFQE LVLACLRSLH S RCIAQEVP KTEMKTKYEW KEDWNLSKPI RIDKAGSDLK AYKTNYNAIF KQRLKQEHIN EQLKQLDNSD FAKQLKIFKP FK IN UniProtKB: Uncharacterized protein |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Concentration | 2.2 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm Bright-field microscopy / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 45.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT | ||||||
| Output model |  PDB-8v3l: |
 Movie
Movie Controller
Controller



 Z
Z Y
Y X
X